-
Notifications
You must be signed in to change notification settings - Fork 85
Description
This issue is split from #407 since the symptom is similar but the underlying cause is different.
Tagging @oftfrfbf, @lsetiawan @emiliom so that we can continue the discussion here. 😃
This issue focuses on the problem that there is sometimes very large expansion of memory use when echopype is converting a file of moderate size.
(vs #407 focuses on the problem when the data file itself is too large to fit into memory.)
Description
Based on @oftfrfbf and @lsetiawan 's investigations (quoted below), there may very well be two different types of things happening (or more? depending on the exact form of data):
-
for this 725 MB OOI file:
- 1 of the 3 channels is split-beam and produces
alongship_angleandathwartship_angledata, but the other 2 single-beam channels do not produce this data. - The data are first parsed into lists and then organized/merged into xarray DataArray/Dataset. During the second stage @lsetiawan pointed out that there is an expansion of memory.
- I suspect this is due to trying to pad NaNs to the (correctly) missing angle data slices in order to assemble a data cube with dimensions
frequency(channel),range_binandping_time.
- 1 of the 3 channels is split-beam and produces
-
for the 95 MB file shared by @oftfrfbf:
Below is @oftfrfbf's profiling result (quoted from here):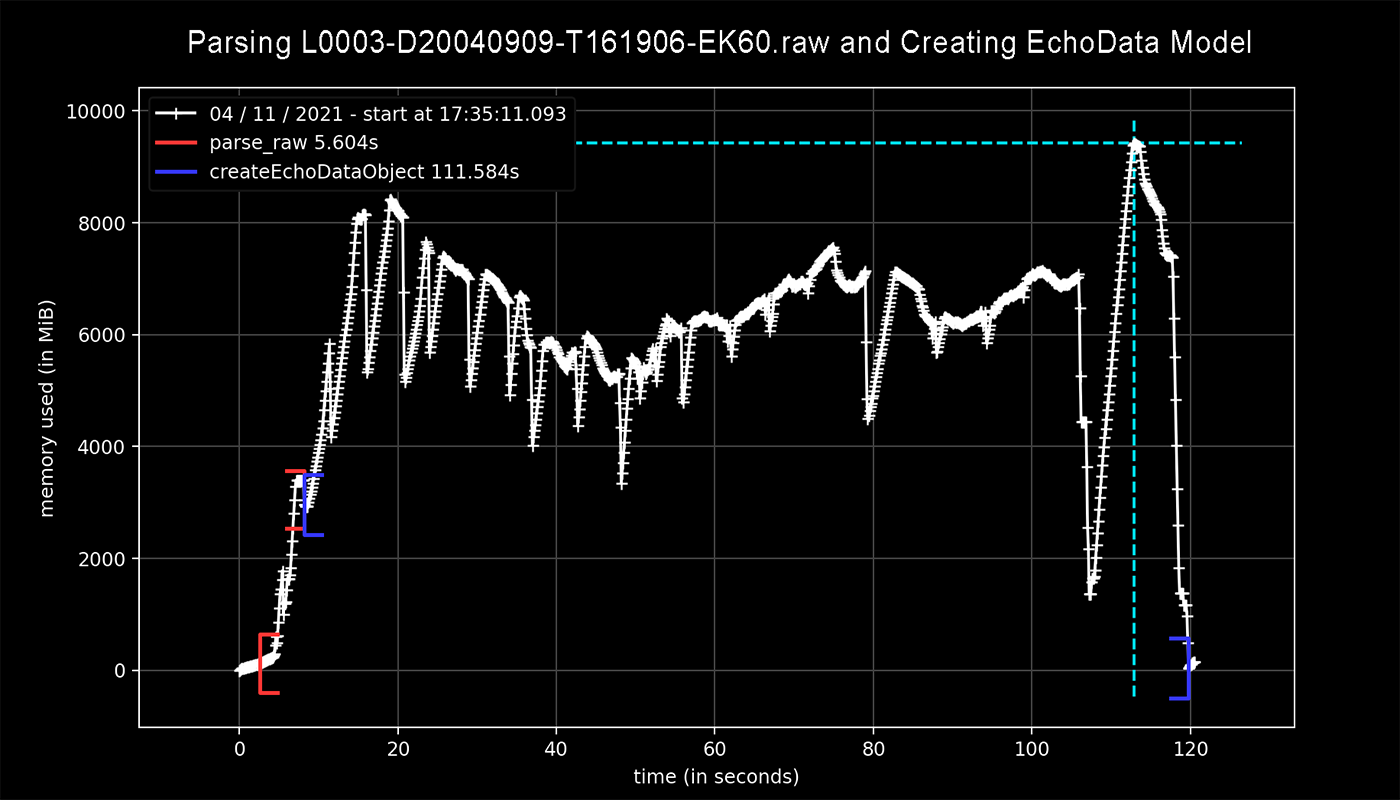
Profiling for parser.parse_raw() (shown in red) only uses about 3 GB of memory while lines 424 through 445, accounting for the creation of the EchoData model (shown in blue), brought that value up to a total of 10 GB.
- I wonder whether the recorded range within this file was changed dramatically between ping to ping? I haven't had a change to look into the file but will try to do so soon and report back.
- The reason why I suspect this is again due to the padding NaN approach to bring pings of different lengths into a 3D data cube: if there are some pings that are somehow much much "longer" than the other pings, all the other pings would be padded with NaN to that longer length, exploding the memory.
Potential solution?
For the case with the OOI file, I wonder if we could circumvent the problem by saving the larger data variables (backscatter_r, alonghip_angle, athwartship_angle) one by one in xr.Dataset.to_zarr using mode='a', so that the max memory usage would not surpass what would be required for one of them (vs having all 3 in memory at the same time).
The case with the 95 MB file is more complicated if the reason is as described above.
A couple of not particularly well-formed thoughts:
- use
.to_zarrwithregionspecification while organizing and storing the Dataset, so that the in-memory presence is smaller. - somehow use sparse arrays like what's discussed here and
pydata/sparsefor the in memory component. There's some discussion on dask support for sparse array here.
Thoughts???
Metadata
Metadata
Assignees
Labels
Type
Projects
Status