Closed
Description
Using pm.Bound for distributions whose arguments include other model parameters (instead of constants) will lead to a model that is wrongly sampled in sample_prior_predictive()
with pm.Model() as m:
pop = pm.Normal('pop', 2, 1)
ind = pm.Bound(pm.Normal, lower=-2, upper=2)('ind', mu=pop, sigma=.5)
prior = pm.sample_prior_predictive()
trace = pm.sample()The samples for the bounded distribution are fine (i.e., one obtains the same results if doing prior_predictive or sampling without data):
sns.kdeplot(prior['ind'])
sns.kdeplot(trace['ind'])But the samples for the hyper-parameters do not match:
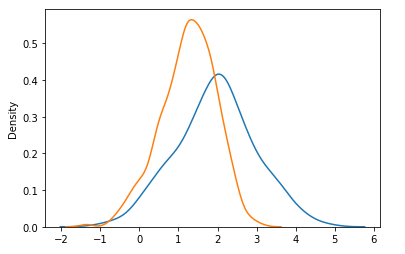
sns.kdeplot(prior['pop'])
sns.kdeplot(trace['pop'])What is going on? I think the way pm.Bound is implemented, it corresponds to adding an arbitrary factor to the model logp of the kind:
pm.Potential('err', pm.math.switch((ind >= -2) & (ind <= 2), 0, -np.inf))Which obviously is not (and cannot) be accounted for in predictive sampling.