diff --git a/.github/workflows/build-and-test-callable.yml b/.github/workflows/build-and-test-callable.yml
new file mode 100644
index 000000000..aac512186
--- /dev/null
+++ b/.github/workflows/build-and-test-callable.yml
@@ -0,0 +1,188 @@
+# build-and-test-callable.yml
+#
+# - Builds PyMFEM in three stages:
+# - MFEM dependencies + MFEM
+# - SWIG bindings
+# - PyMFEM
+# - Runs tests under `run_examples.py`
+# - If there is a failure, uploads test outputs as an artifact
+
+name: Build and Test
+
+on:
+ workflow_call:
+ inputs:
+ os:
+ description: 'Operating system'
+ type: string
+ default: 'ubuntu-latest'
+ python-version:
+ description: 'Python version'
+ type: string
+ default: '3.9'
+ mfem-branch:
+ description: 'MFEM branch to checkout'
+ type: string
+ default: 'default' # 'default' uses a specific commit hash defined in setup.py:repos_sha
+ parallel:
+ description: 'Build parallel version'
+ type: boolean
+ default: false
+ cuda:
+ description: 'Build with CUDA'
+ type: boolean
+ default: false
+ cuda-toolkit-version:
+ type: string
+ default: '12.6.0'
+ cuda-driver-version:
+ type: string
+ default: '560.28.03'
+ libceed:
+ description: 'Build with libCEED'
+ type: boolean
+ default: false
+ gslib:
+ description: 'Build with GSlib'
+ type: boolean
+ default: false
+ phases:
+ description: 'When true, run each build step individually (mfem, swig, pymfem)'
+ type: boolean
+ default: true
+
+jobs:
+ build-and-test:
+ runs-on: ${{ inputs.os }}
+
+ # Reference for $${{ x && y || z }} syntax: https://7tonshark.com/posts/github-actions-ternary-operator/
+ name: >-
+ ${{ inputs.os }} |
+ ${{ inputs.mfem-branch }} |
+ ${{ inputs.python-version }} |
+ ${{ inputs.parallel && 'parallel' || 'serial' }}
+ ${{ inputs.cuda && '| cuda' || '' }}${{ inputs.libceed && '| libceed' || '' }}${{ inputs.gslib && '| gslib' || '' }}
+
+ env:
+ CUDA_HOME: '/usr/local/cuda'
+ # These are all passed to setup.py as one concatenated string
+ build-flags: >-
+ ${{ inputs.parallel && '--with-parallel' || '' }}
+ ${{ inputs.cuda && '--with-cuda' || '' }}
+ ${{ inputs.libceed && '--with-libceed' || '' }}
+ ${{ inputs.gslib && '--with-gslib' || '' }}
+ ${{ (!(inputs.mfem-branch == 'default') && format('--mfem-branch=''{0}''', inputs.mfem-branch)) || '' }}
+
+ # -------------------------------------------------------------------------------------------------
+ # Begin workflow
+ # -------------------------------------------------------------------------------------------------
+ steps:
+ - name: Checkout repo
+ uses: actions/checkout@v4
+
+ - name: Set up Python ${{ inputs.python-version }}
+ uses: actions/setup-python@v5
+ with:
+ python-version: ${{ inputs.python-version }}
+
+ # -------------------------------------------------------------------------------------------------
+ # Download/install dependencies
+ # -------------------------------------------------------------------------------------------------
+ - name: Install core dependencies via requirements.txt
+ run: |
+ pip install setuptools
+ pip install -r requirements.txt --verbose
+
+ - name: Install MPI
+ if: inputs.parallel
+ run: |
+ sudo apt-get install openmpi-bin libopenmpi-dev
+ export OMPI_MCA_rmaps_base_oversubscribe=1
+ # (broken?) sudo apt-get install mpich libmpich-dev
+ pip install mpi4py
+ mpirun -np 2 python -c "from mpi4py import MPI;print(MPI.COMM_WORLD.rank)"
+
+ - name: Purge PIP chach
+ run: pip cache purge
+
+ - name: Cache CUDA
+ if: inputs.cuda
+ id: cache-cuda
+ uses: actions/cache@v4
+ with:
+ path: ~/cache
+ key: cuda-installer-${{ inputs.cuda-toolkit-version }}-${{ inputs.cuda-driver-version }}
+
+ - name: Download CUDA
+ if: inputs.cuda && steps.cache-cuda.outputs.cache-hit == false
+ run: |
+ CUDA_URL="https://developer.download.nvidia.com/compute/cuda/${{ inputs.cuda-toolkit-version }}/local_installers/cuda_${{ inputs.cuda-toolkit-version }}_${{ inputs.cuda-driver-version }}_linux.run"
+ curl -o ~/cache/cuda.run --create-dirs $CUDA_URL
+
+ - name: Install CUDA
+ if: inputs.cuda
+ run: |
+ # The --silent flag is necessary to bypass user-input, e.g. accepting the EULA
+ sudo sh ~/cache/cuda.run --silent --toolkit
+ echo "/usr/local/cuda/bin" >> $GITHUB_PATH
+
+ - name: Print dependency information
+ run: |
+ pip list
+ printf "\n\n---------- MPI ----------\n"
+ mpiexec --version || printf "MPI not installed"
+ printf "\n\n---------- CUDA ----------\n"
+ nvcc --version || printf "CUDA not installed"
+
+ # -------------------------------------------------------------------------------------------------
+ # Build MFEM + SWIG Bindings + PyMFEM
+ # -------------------------------------------------------------------------------------------------
+ - name: Build MFEM (step 1)
+ if: inputs.phases
+ run: python setup.py install --ext-only --vv ${{ env.build-flags }}
+
+ - name: Build SWIG wrappers (step 2)
+ if: inputs.phases
+ run: python setup.py install --swig --vv ${{ env.build-flags }}
+
+ - name: Build PyMFEM (step 3)
+ if: inputs.phases
+ run: python setup.py install --skip-ext --skip-swig --vv ${{ env.build-flags }}
+
+ - name: Build all (steps 1-3)
+ if: inputs.phases == false
+ run: python setup.py install --vv ${{ env.build-flags }}
+
+ # -------------------------------------------------------------------------------------------------
+ # Run tests
+ # -------------------------------------------------------------------------------------------------
+ - name: Run tests (serial)
+ if: inputs.parallel == false
+ run: |
+ cd test
+ python run_examples.py -serial -verbose
+
+ - name: Run tests (parallel)
+ if: inputs.parallel
+ run: |
+ cd test
+ python run_examples.py -parallel -verbose -np 2
+
+ # -------------------------------------------------------------------------------------------------
+ # Generate an artifact (output of tests) on failure
+ # -------------------------------------------------------------------------------------------------
+ - name: Generate test results artifact
+ id: generate-artifact
+ run: |
+ tar -cvzf sandbox.tar.gz test/sandbox
+ # generate a name for the artifact
+ txt=$(python -c "import datetime;print(datetime.datetime.now().strftime('%H_%M_%S_%f'))")
+ echo name="test_results_"${txt}"_"${{ github.run_id }}".tar.gz" >> $GITHUB_OUTPUT
+
+ - name: Upload Artifact
+ uses: actions/upload-artifact@v4
+ if: failure()
+ with:
+ name: ${{ steps.generate-artifact.outputs.name }}
+ path: sandbox.tar.gz
+ retention-days: 1
diff --git a/.github/workflows/build-and-test-dispatch.yml b/.github/workflows/build-and-test-dispatch.yml
new file mode 100644
index 000000000..6f0ff98db
--- /dev/null
+++ b/.github/workflows/build-and-test-dispatch.yml
@@ -0,0 +1,119 @@
+# build-and-test-dispatch.yml
+#
+# Dispatch workflow for build-and-test-callable.yml
+name: Build and Test (dispatch)
+
+on:
+ workflow_dispatch:
+ inputs:
+ test_options:
+ type: choice
+ required: true
+ description: 'Test all options. If set to false, will only trigger the CI for the default options.'
+ default: 'defaults'
+ options:
+ - 'defaults'
+ - 'fast'
+ - 'cuda'
+ - 'libceed'
+ - 'gslib'
+ - 'all'
+ pull_request:
+
+jobs:
+ # -------------------------------------------------------------------------------------------------
+ # Build and test whole matrix of options on linux
+ # -------------------------------------------------------------------------------------------------
+ test-linux-serial:
+ if: ${{ github.event_name == 'pull_request' && !contains(github.event.pull_request.labels.*.name, 'skip-ci') || inputs.test_options == 'all' }}
+ strategy:
+ fail-fast: false
+ matrix:
+ mfem-branch: [master, default] # 'default' uses a specific commit hash defined in setup.py:repos_sha
+ python-version: ['3.8', '3.9', '3.10', '3.11', '3.12'] # 3.12 is not supported by scipy
+ parallel: [false]
+ name: test-linux | ${{ matrix.mfem-branch }} | ${{ matrix.python-version }} | ${{ matrix.parallel && 'parallel' || 'serial' }}
+ uses: ./.github/workflows/build-and-test-callable.yml
+ with:
+ os: ubuntu-latest
+ mfem-branch: ${{ matrix.mfem-branch }}
+ python-version: ${{ matrix.python-version }}
+ parallel: ${{ matrix.parallel }}
+
+ test-linux-parallel:
+ if: ${{ github.event_name == 'pull_request' && !contains(github.event.pull_request.labels.*.name, 'skip-ci') || inputs.test_options == 'all' }}
+ strategy:
+ fail-fast: false
+ matrix:
+ mfem-branch: [master, default] # 'default' uses a specific commit hash defined in setup.py:repos_sha
+ python-version: ['3.9', '3.10', '3.11', '3.12'] # 3.12 is not supported by scipy
+ parallel: [true]
+ name: test-linux | ${{ matrix.mfem-branch }} | ${{ matrix.python-version }} | ${{ matrix.parallel && 'parallel' || 'serial' }}
+ uses: ./.github/workflows/build-and-test-callable.yml
+ with:
+ os: ubuntu-latest
+ mfem-branch: ${{ matrix.mfem-branch }}
+ python-version: ${{ matrix.python-version }}
+ parallel: ${{ matrix.parallel }}
+
+ # -------------------------------------------------------------------------------------------------
+ # Fast test
+ # -------------------------------------------------------------------------------------------------
+ test-fast:
+ if: ${{ inputs.test_options == 'fast' }}
+ strategy:
+ fail-fast: false
+ matrix:
+ mfem-branch: [master, default] # 'default' uses a specific commit hash defined in setup.py:repos_sha
+ parallel: [false, true]
+ name: test-fast | ${{ matrix.mfem-branch }} | ${{ matrix.parallel && 'parallel' || 'serial' }}
+ uses: ./.github/workflows/build-and-test-callable.yml
+ with:
+ os: ubuntu-latest
+ mfem-branch: ${{ matrix.mfem-branch }}
+ python-version: '3.9'
+ parallel: ${{ matrix.parallel }}
+
+ # -------------------------------------------------------------------------------------------------
+ # Specific cases (we want these to use defaults, and not expand the dimensions of the matrix)
+ # -------------------------------------------------------------------------------------------------
+ test-macos:
+ if: ${{ github.event_name == 'pull_request' && !contains(github.event.pull_request.labels.*.name, 'skip-ci') || inputs.test_options == 'all' }}
+ strategy:
+ fail-fast: false
+ matrix:
+ mfem-branch: [master, default]
+ name: test-macos | ${{ matrix.mfem-branch }}
+ uses: ./.github/workflows/build-and-test-callable.yml
+ with:
+ os: macos-latest
+ mfem-branch: ${{ matrix.mfem-branch }}
+
+ test-cuda:
+ if: ${{ github.event_name == 'pull_request' && !contains(github.event.pull_request.labels.*.name, 'skip-ci') || inputs.test_options == 'all' || inputs.test_options == 'cuda'}}
+ uses: ./.github/workflows/build-and-test-callable.yml
+ with:
+ cuda: true
+ name: test-cuda
+
+ test-libceed:
+ if: ${{ github.event_name == 'pull_request' && !contains(github.event.pull_request.labels.*.name, 'skip-ci') || inputs.test_options == 'all' || inputs.test_options == 'libceed'}}
+ uses: ./.github/workflows/build-and-test-callable.yml
+ with:
+ libceed: true
+ name: test-libceed
+
+ test-gslib:
+ if: ${{ github.event_name == 'pull_request' && !contains(github.event.pull_request.labels.*.name, 'skip-ci') || inputs.test_options == 'all' || inputs.test_options == 'gslib'}}
+ uses: ./.github/workflows/build-and-test-callable.yml
+ with:
+ gslib: true
+ name: test-gslib
+
+ # -------------------------------------------------------------------------------------------------
+ # Build and test defaults
+ # -------------------------------------------------------------------------------------------------
+ test-defaults:
+ if: ${{ github.event_name == 'workflow_dispatch' && inputs.test_options == 'defaults' }}
+ uses: ./.github/workflows/build-and-test-callable.yml
+ name: test-defaults
diff --git a/.github/workflows/release_binary.yml b/.github/workflows/release_binary.yml
index 0437a2473..df79b5e92 100644
--- a/.github/workflows/release_binary.yml
+++ b/.github/workflows/release_binary.yml
@@ -36,11 +36,11 @@ jobs:
ls -l dist/
#python3 -m twine upload --repository-url https://test.pypi.org/legacy/ --password ${{ secrets.TEST_PYPI_TOKEN }} --username __token__ --verbose dist/*
python3 -m twine upload --password ${{ secrets.PYPI_TOKEN }} --username __token__ --verbose dist/*
- make_binary_3_7_8_9_10:
+ make_binary_3_8_9_10_11:
needs: make_sdist
strategy:
matrix:
- pythonpath: ["cp37-cp37m", "cp38-cp38", "cp39-cp39", "cp310-cp310"]
+ pythonpath: ["cp38-cp38", "cp39-cp39", "cp310-cp310", "cp311-cp311"]
runs-on: ubuntu-latest
container: quay.io/pypa/manylinux2014_x86_64
@@ -59,14 +59,10 @@ jobs:
#
ls -l /opt/python/
export PATH=/opt/python/${{ matrix.pythonpath }}/bin:$PATH
- pip3 install six auditwheel twine
+ pip3 install auditwheel twine
pip3 install urllib3==1.26.6 # use older version to avoid OpenSSL vesion issue
- pip3 install numpy==1.21.6
- pip3 install numba-scipy
- if [ -f requirements.txt ]; then
- pip3 install -r requirements.txt
- fi
+ pip3 install -r requirements.txt
CWD=$PWD
yum install -y zlib-devel
diff --git a/.github/workflows/test_with_MFEM_master.yml b/.github/workflows/test_with_MFEM_master.yml
deleted file mode 100644
index a2c3ca0c0..000000000
--- a/.github/workflows/test_with_MFEM_master.yml
+++ /dev/null
@@ -1,157 +0,0 @@
-name: Test_with_MFEM_master
-
-on:
- pull_request:
- types:
- - labeled
- #push:
- # branches: [ test ]
- #pull_request:
- # branches: [ test ]
- # types: [synchronize]
-
-jobs:
- build:
- if: contains(github.event.pull_request.labels.*.name, 'in-test-with-mfem-master')
- strategy:
- fail-fast: false
- matrix:
- python-version: ["3.7", "3.8", "3.9", "3.10"]
- #python-version: ["3.10"]
- os: [ubuntu-20.04]
- # USE_FLAGS : cuda, parallel, libceed
- env:
- - { USE_FLAGS: "000"}
- - { USE_FLAGS: "100"}
- - { USE_FLAGS: "010"}
- - { USE_FLAGS: "110"}
-
- include:
- - os: macos-latest
- python-version: 3.9
- env: {USE_FLAGS: "000"}
-# - os: [ubuntu-20.04]
-# python-version: 3.9
-# env: {USE_FLAGS: "001"}
-#
- runs-on: ${{ matrix.os }}
- #env: ${{ matrix.env }}
- env:
- USE_FLAGS: ${{ matrix.env.USE_FLAGS }}
- CUDA: "11.5"
-
- steps:
- - uses: actions/checkout@v3
-# with:
-# ref: master_tracking_branch
-
- - name: Set up Python ${{ matrix.python-version }}
- uses: actions/setup-python@v4
- with:
- python-version: ${{ matrix.python-version }}
-
- - name: Install dependencies
- run: |
- #python -m pip install --upgrade pip setuptools wheel
- python -c "import setuptools;print(setuptools.__version__)"
-
- PYTHONLIB=${HOME}/sandbox/lib/python${{ matrix.python-version }}/site-packages
- mkdir -p $PYTHONLIB
-
- export PYTHONPATH=$PYTHONLIB:$PYTHONPATH
- echo "PYTHONPATH:"$PYTHONPATH
-
- pip install six numpy --verbose
- if [ "${{matrix.python-version}}" = "3.10" ] ; then
- pip install numba numba-scipy --verbose
- #pip install scipy numba --verbose
- else
- pip install numba numba-scipy --verbose
- #pip install numba scipy --verbose
- fi
-
- if [ -f requirements.txt ]; then
- pip install -r requirements.txt --prefix=${HOME}/sandbox --verbose
- fi
-
- python -c "import sys;print(sys.path)"
- python -c "import numpy;print(numpy.__file__)"
-
- echo ${HOME}
- which swig # this default is 4.0.1
- pip install swig --prefix=${HOME}/sandbox
- export PATH=/usr/local/cuda-${CUDA}/bin:${HOME}/sandbox/bin:$PATH
- ls ${HOME}/sandbox/bin
- which swig
- swig -version
- ${HOME}/sandbox/bin/swig -version
-
- if [ "${USE_FLAGS:0:1}" = "1" ] ; then
- echo $cuda
- source ./ci_scripts/add_cuda_11_5.sh;
- fi
- if [ "${USE_FLAGS:1:1}" = "1" ] ; then
- sudo apt-get install mpich;
- sudo apt-get install libmpich-dev;
- pip install mpi4py
- #pip install mpi4py --no-cache-dir --prefix=${HOME}/sandbox;
- #python -m pip install git+https://github.com/mpi4py/mpi4py
- #python -c "import mpi4py;print(mpi4py.get_include())";
- fi
- ls -l $PYTHONLIB
-
- - name: Build
- run: |
- export PATH=/usr/local/cuda-${CUDA}/bin:${HOME}/sandbox/bin:$PATH
- PYTHONLIB=${HOME}/sandbox/lib/python${{ matrix.python-version }}/site-packages
- export PYTHONPATH=$PYTHONLIB:$PYTHONPATH
- echo $PATH
- echo $PYTHONPATH
- echo "SWIG exe"$(which swig)
-
- if [ "${USE_FLAGS}" = "000" ]; then
- python setup.py install --user --mfem-branch='master'
- fi
- if [ "${USE_FLAGS}" = "010" ]; then
- python setup.py install --with-parallel --prefix=${HOME}/sandbox --mfem-branch='master'
- fi
- if [ "${USE_FLAGS}" = "100" ]; then
- ls -l /usr/local/cuda-${CUDA}/include
- python setup.py install --with-cuda --prefix=${HOME}/sandbox --mfem-branch='master'
- fi
- if [ "${USE_FLAGS}" = "110" ]; then
- python setup.py install --with-cuda --with-parallel --mfem-branch='master'
- fi
- if [ "${USE_FLAGS}" = "001" ]; then
- python setup.py install --prefix=${HOME}/sandbox --with-libceed --mfem-branch='master'
- fi
-
- - name: RUN_EXAMPLES
- run: |
- export PATH=/usr/local/cuda-${CUDA}/bin:$PATH
- PYTHONLIB=${HOME}/sandbox/lib/python${{ matrix.python-version }}/site-packages
- export PYTHONPATH=$PYTHONLIB:$PYTHONPATH
- echo $PATH
- echo $PYTHONPATH
- cd test
- echo $PATH
- echo $PYTHONPATH
- if [ "${USE_FLAGS}" = "000" ]; then
- #python ../examples/ex1.py
- #python ../examples/ex9.py
- python run_examples.py -serial -verbose
- fi
- if [ "${USE_FLAGS}" = "010" ]; then
- which mpicc
- python run_examples.py -parallel -verbose -np 2
- fi
- if [ "${USE_FLAGS}" = "100" ]; then
- python ../examples/ex1.py --pa
- fi
- if [ "${USE_FLAGS}" = "110" ]; then
- mpirun -np 2 python ../examples/ex1.py --pa
- fi
- if [ "${USE_FLAGS}" = "001" ]; then
- python ../examples/ex1.py -d ceed-cpu
- python ../examples/ex9.py -d ceed-cpu
- fi
diff --git a/.github/workflows/test_with_MFEM_release.yml b/.github/workflows/test_with_MFEM_release.yml
deleted file mode 100644
index 9d5e6db1d..000000000
--- a/.github/workflows/test_with_MFEM_release.yml
+++ /dev/null
@@ -1,158 +0,0 @@
-name: Test_with_MFEM_release
-
-on:
- pull_request:
- types:
- - labeled
- #push:
- # branches: [ test ]
- #pull_request:
- # branches: [ test ]
- # types: [synchronize]
-
-jobs:
- build:
- if: contains(github.event.pull_request.labels.*.name, 'in-test-with-mfem-release')
- strategy:
- fail-fast: false
- matrix:
- python-version: ["3.7", "3.8", "3.9"]
- #python-version: ["3.10"]
- #os: [ubuntu-latest]
- os: [ubuntu-20.04]
- # USE_FLAGS : cuda, parallel, libceed
- env:
- - { USE_FLAGS: "000"}
- - { USE_FLAGS: "010"}
-
- include:
- - os: macos-latest
- python-version: 3.9
- env: {USE_FLAGS: "000"}
-
- runs-on: ${{ matrix.os }}
- #env: ${{ matrix.env }}
- env:
- USE_FLAGS: ${{ matrix.env.USE_FLAGS }}
- CUDA: "11.5"
- SANDBOX: ~/sandbox
-
- steps:
- - uses: actions/checkout@v3
-# with:
-# ref: master_tracking_branch
-
- - name: Set up Python ${{ matrix.python-version }}
- uses: actions/setup-python@v4
- with:
- python-version: ${{ matrix.python-version }}
-
- - name: Install dependencies
- run: |
- #python -m pip install --upgrade pip setuptools wheel
- python -c "import setuptools;print(setuptools.__version__)"
-
- PYTHONLIB=~/sandbox/lib/python${{ matrix.python-version }}/site-packages
- mkdir -p $PYTHONLIB
-
- export PYTHONPATH=$PYTHONLIB:$PYTHONPATH
- echo "PYTHONPATH:"$PYTHONPATH
-
- pip install six --verbose
- if [ "${{matrix.python-version}}" = "3.10" ] ; then
- pip install numba numba-scipy --verbose
- #pip install scipy
- else
- pip install numba numba-scipy --verbose
- fi
-
- if [ -f requirements.txt ]; then
- pip install -r requirements.txt --prefix=~/sandbox --verbose
- fi
-
- python -c "import sys;print(sys.path)"
- python -c "import numpy;print(numpy.__file__)"
-
- which swig # this default is 4.0.1
- pip install swig --prefix=~/sandbox
- export PATH=/usr/local/cuda-${CUDA}/bin:~/sandbox/bin:$PATH
- which swig
- swig -version
-
- if [ "${USE_FLAGS:0:1}" = "1" ] ; then
- echo $cuda
- source ./ci_scripts/add_cuda_11_5.sh;
- fi
- if [ "${USE_FLAGS:1:1}" = "1" ] ; then
- sudo apt-get install mpich;
- sudo apt-get install libmpich-dev;
- pip install mpi4py --prefix=~/sandbox
- #pip install mpi4py --no-cache-dir --prefix=~/sandbox;
- #python -m pip install git+https://github.com/mpi4py/mpi4py
- python -c "import mpi4py;print(mpi4py.get_include())";
- fi
- ls -l $PYTHONLIB
-
- - name: Build
- run: |
- export PATH=/usr/local/cuda-${CUDA}/bin:~/sandbox/bin:$PATH
- PYTHONLIB=~/sandbox/lib/python${{ matrix.python-version }}/site-packages
- export PYTHONPATH=$PYTHONLIB:$PYTHONPATH
- echo $PATH
- echo $PYTHONPATH
- echo $SANDBOX
- echo "SWIG exe"$(which swig)
-
- if [ "${USE_FLAGS}" = "000" ]; then
- # test workflow to manually run swig
- python setup.py clean --swig
- python setup.py install --ext-only --with-gslib --verbose
- python setup.py install --swig --with-gslib --verbose
- python setup.py install --skip-ext --skip-swig --with-gslib --verbose
- fi
- if [ "${USE_FLAGS}" = "010" ]; then
- python setup.py install --with-gslib --with-parallel --prefix=$SANDBOX
- fi
-
- - name: RUN_EXAMPLES
- run: |
-
- export PATH=/usr/local/cuda-${CUDA}/bin:$PATH
- PYTHONLIB=~/sandbox/lib/python${{ matrix.python-version }}/site-packages
- export PYTHONPATH=$PYTHONLIB:$PYTHONPATH
- echo $PATH
- echo $PYTHONPATH
- echo $SANDBOX
- cd test
- echo $PATH
- echo $PYTHONPATH
- if [ "${USE_FLAGS}" = "000" ]; then
- python run_examples.py -serial -verbose
- fi
- if [ "${USE_FLAGS}" = "010" ]; then
- which mpicc
- python run_examples.py -parallel -verbose -np 2
- fi
-
- - name: Generate Artifact
- if: always()
- run: |
- tar -cvzf sandbox.tar.gz test/sandbox
-
- - name: Generate artifact name
- if: always()
- id: generate-name
- run: |
- txt=$(python -c "import datetime;print(datetime.datetime.now().strftime('%H_%M_%S_%f'))")
- name="test_result_"${txt}"_"${{ github.run_id }}".tar.gz"
- echo $name
- # (deprecated) echo "::set-output name=artifact::${name}"
- echo "artifact=${name}" >> $GITHUB_OUTPUT
-
- - name: Upload Artifact
- uses: actions/upload-artifact@v3
- if: failure()
- with:
- name: ${{ steps.generate-name.outputs.artifact }}
- path: sandbox.tar.gz
- retention-days: 1
diff --git a/.github/workflows/testrelease_binary.yml b/.github/workflows/testrelease_binary.yml
index 15b099053..ae08741eb 100644
--- a/.github/workflows/testrelease_binary.yml
+++ b/.github/workflows/testrelease_binary.yml
@@ -36,11 +36,11 @@ jobs:
ls -l dist/
python3 -m twine upload --repository-url https://test.pypi.org/legacy/ --password ${{ secrets.TEST_PYPI_TOKEN }} --username __token__ --verbose dist/*
#python3 -m twine upload --password ${{ secrets.PYPI_TOKEN }} --username __token__ --verbose dist/*
- make_binary_3_7_8_9_10:
+ make_binary_3_8_9_10_11:
needs: make_sdist
strategy:
matrix:
- pythonpath: ["cp37-cp37m", "cp38-cp38", "cp39-cp39", "cp310-cp310"]
+ pythonpath: ["cp38-cp38", "cp39-cp39", "cp310-cp310", "cp311-cp311"]
runs-on: ubuntu-latest
container: quay.io/pypa/manylinux2014_x86_64
@@ -59,14 +59,11 @@ jobs:
#
ls -l /opt/python/
export PATH=/opt/python/${{ matrix.pythonpath }}/bin:$PATH
- pip3 install six auditwheel twine
- pip3 install numpy==1.21.6
+ pip3 install auditwheel twine
pip3 install urllib3==1.26.6 # use older version to avoid OpenSSL vesion issue
- pip3 install numba-scipy
- if [ -f requirements.txt ]; then
- pip3 install -r requirements.txt
- fi
+
+ pip3 install -r requirements.txt
CWD=$PWD
yum install -y zlib-devel
diff --git a/INSTALL.md b/INSTALL.md
new file mode 100644
index 000000000..dcd61d519
--- /dev/null
+++ b/INSTALL.md
@@ -0,0 +1,178 @@
+# Installation Guide
+
+## Basic install
+
+Most users will be fine using the binary bundled in the default `pip` install:
+
+```shell
+pip install mfem
+```
+The above installation will download and install a *serial* version of `MFEM`.
+
+## Building from source
+PyMFEM has many options for installation, when building from source, including:
+ - Serial and parallel (MPI) wrappers
+ - Using pre-built local dependencies
+ - Installing additional dependencies such as
+ - `hypre`
+ - `gslib`
+ - `libceed`
+ - `metis`
+
+Most of the options for PyMFEM can be used directly when installing via `python setup.py install`, e.g.
+```shell
+git clone git@github:mfem/PyMFEM.git
+cd PyMFEM
+python setup.py install --user
+```
+For example, parallel (MPI) support is built with the `--with-parallel` flag:
+```shell
+python setup.py install --with-parallel
+```
+
+Note: this option turns on building `metis` and `Hypre`
+
+## Commonly used flags
+
+| Flag | Description |
+|------|-------------|
+| `--with-parallel` | Install both serial and parallel versions of `MFEM` and the wrapper
(note: this option turns on building `metis` and `hypre`) |
+| `--mfem-branch=` | Download/install MFEM using a specific reference (`git` `branch`, `hash`, or `tag`) |
+| `--user` | Install in user's site-package |
+
+In order to see the full list of options, use
+
+```shell
+python setup.py install --help
+```
+
+## Advanced options
+
+### Suitesparse
+`--with-suitesparse` : build MFEM with `suitesparse`. `suitesparse` needs to be installed separately.
+Point to the location of `suitesparse` using the flag `--suitesparse-prefix=`
+
+Note: this option turns on building `metis` in serial
+
+### CUDA
+`--with-cuda` : build MFEM with CUDA. Hypre cuda build is also supported using
+`--with-cuda-hypre`. `--cuda-arch` can be used to specify cuda compute capablility.
+(See table in https://en.wikipedia.org/wiki/CUDA#Supported_GPUs)
+
+CUDA needs to be installed separately and nvcc must be found in PATH ([Example](https://github.com/mfem/PyMFEM/blob/e1466a6a/.github/workflows/build-and-test-callable.yml#L111-L122)).
+
+(examples)
+```shell
+python setup.py install --with-cuda
+
+python setup.py install --with-cuda --with-cuda-hypre
+
+python setup.py install --with-cuda --with-cuda-hypre --cuda-arch=80 (A100)
+
+python setup.py install --with-cuda --with-cuda-hypre --cuda-arch=75 (Turing)
+```
+
+### gslib
+`--with-gslib` : build MFEM with [GSlib](https://github.com/Nek5000/gslib)
+
+Note: this option downloads and builds GSlib
+
+### libCEED
+`--with-libceed` : build MFEM with [libCEED](https://github.com/CEED/libCEED)
+
+Note: this option downloads and builds libCEED
+
+### Specifying compilers
+| Flag | Description |
+|------|--------|
+| `--CC` | c compiler |
+| `--CXX` | c++ compiler |
+| `--MPICC` | mpic compiler |
+| `--MPICXX` | mpic++ compiler |
+
+(example)
+Using Intel compiler
+```shell
+python setup.py install --with-parallel --CC=icc, --CXX=icpc, --MPICC=mpiicc, --MPICXX=mpiicpc
+```
+
+### Building MFEM with specific version
+By default, setup.py build MFEM with specific SHA (which is usually the released latest version).
+In order to use the latest MFEM in Github. One can specify the branch name or SHA using mfem-branch
+option.
+
+`--mfem-branch = `
+
+(example)
+```shell
+python setup.py install --mfem-branch=master
+```
+
+### Using MFEM build externally.
+These options are used to link PyMFEM wrapper with existing MFEM library. We need `--mfem-source`
+and `--mfem-prefix`
+
+| Flag | Description |
+|----------------------------|-------------------------------------------------------------------|
+| `--mfem-source ` | The location of MFEM source used to build MFEM |
+| `--mfem-prefix ` | The location of the MFEM library. `libmfem.so` needs to be found in `/lib` |
+| `--mfems-prefix `| (optional) Specify serial MFEM location separately |
+| `--mfemp-prefix `| (optional) Specify parallel MFEM location separately |
+
+
+### Blas and Lapack
+--with-lapack : build MFEM with lapack
+
+`` is used for CMAKE call to buid MFEM
+`--blas-libraries=`
+`--lapack-libraries=`
+
+### Options for development and testing
+| Flag | Description |
+|------|--------|
+| `--swig` | run swig only |
+| `--skip-swig` | build without running swig` |
+| `--skip-ext` | skip building external libraries.|
+| `--ext-only` | build exteranl libraries and exit.|
+
+During the development, often we update depenencies (such as MFEM) and edit `*.i` file.
+
+
+First clean everything.
+
+```shell
+python setup.py clean --all
+```
+
+Then, build externals alone
+```shell
+python setup.py install --with-parallel --ext-only --mfem-branch=master
+```
+
+Then, genrate swig wrappers.
+```shell
+python setup.py install --with-parallel --swig --mfem-branch=master
+```
+
+If you are not happy with the wrapper (`*.cxx` and `*.py`), you edit `*.i` and redo
+the same. When you are happy, build the wrapper. `--swig` does not clean the
+existing wrapper. So, it will only update wrapper for updated `*.i`
+
+When building a wrapper, you can use `--skip-ext` option. By default, it will re-run
+swig to generate entire wrapper codes.
+```shell
+python setup.py install --with-parallel --skip-ext --mfem-branch=master
+```
+
+If you are sure, you could use `--skip-swig` option, so that it compiles the wrapper
+codes without re-generating it.
+```shell
+python setup.py install --with-parallel --skip-ext --skip-swig --mfem-branch=master
+```
+
+### Other options
+`--unverifiedSSL` :
+ This addresses error relating SSL certificate. Typical error message is
+ ``
+
+
diff --git a/README.md b/README.md
index ef631a977..fe8806552 100644
--- a/README.md
+++ b/README.md
@@ -11,9 +11,8 @@ By default, "pip install mfem" downloads and builds the serial version of MFEM a
Additionally, the installer supports building MFEM with specific options together with other external libraries, including MPI version.
## Install
-```
+```shell
pip install mfem # binary install is available only on linux platforms (Py36-310)
-pip install mfem --no-binary mfem # install serial MFEM + wrapper from source
```
@@ -23,13 +22,9 @@ with pip, or download the package manually and run the script. For example, the
and build parallel version of MFEM library (linked with Metis and Hypre)
and installs under /mfem. See also, docs/install.txt
-### Using pip
-```
-$ pip install mfem --install-option="--with-parallel"
-```
### Build from local source file
-```
+```shell
# Download source and build
$ pip download mfem --no-binary mfem (expand tar.gz file and move to the downloaded directory)
or clone this repository
@@ -59,36 +54,42 @@ $ python setup.py install --help
```
## Usage
-Here is an example to solve div(alpha grad(u)) = f in a square and to plot the result
-with matplotlib (modified from ex1.cpp). Use the badge above to open this in Binder.
-```
+This example (modified from `ex1.cpp`) solves the Poisson equation,
+$$\nabla \cdot (\alpha \nabla u) = f$$
+in a square and plots the result using matplotlib.
+Use the badge above to open this in Binder.
+
+```python
import mfem.ser as mfem
-# create sample mesh for square shape
+# Create a square mesh
mesh = mfem.Mesh(10, 10, "TRIANGLE")
-# create finite element function space
+# Define the finite element function space
fec = mfem.H1_FECollection(1, mesh.Dimension()) # H1 order=1
fespace = mfem.FiniteElementSpace(mesh, fec)
-#
+# Define the essential dofs
ess_tdof_list = mfem.intArray()
ess_bdr = mfem.intArray([1]*mesh.bdr_attributes.Size())
fespace.GetEssentialTrueDofs(ess_bdr, ess_tdof_list)
-# constant coefficient (diffusion coefficient and RHS)
+# Define constants for alpha (diffusion coefficient) and f (RHS)
alpha = mfem.ConstantCoefficient(1.0)
rhs = mfem.ConstantCoefficient(1.0)
-# Note:
-# Diffusion coefficient can be variable. To use numba-JIT compiled
-# functio. Use the following, where x is numpy-like array.
-# @mfem.jit.scalar
-# def alpha(x):
-# return x+1.0
-#
+"""
+Note
+-----
+In order to represent a variable diffusion coefficient, you
+must use a numba-JIT compiled function. For example:
-# define Bilinear and Linear operator
+>>> @mfem.jit.scalar
+>>> def alpha(x):
+>>> return x+1.0
+"""
+
+# Define the bilinear and linear operators
a = mfem.BilinearForm(fespace)
a.AddDomainIntegrator(mfem.DiffusionIntegrator(alpha))
a.Assemble()
@@ -96,38 +97,37 @@ b = mfem.LinearForm(fespace)
b.AddDomainIntegrator(mfem.DomainLFIntegrator(rhs))
b.Assemble()
-# create gridfunction, which is where the solution vector is stored
-x = mfem.GridFunction(fespace);
+# Initialize a gridfunction to store the solution vector
+x = mfem.GridFunction(fespace)
x.Assign(0.0)
-# form linear equation (AX=B)
+# Form the linear system of equations (AX=B)
A = mfem.OperatorPtr()
B = mfem.Vector()
X = mfem.Vector()
-a.FormLinearSystem(ess_tdof_list, x, b, A, X, B);
+a.FormLinearSystem(ess_tdof_list, x, b, A, X, B)
print("Size of linear system: " + str(A.Height()))
-# solve it using PCG solver and store the solution to x
+# Solve the linear system using PCG and store the solution in x
AA = mfem.OperatorHandle2SparseMatrix(A)
M = mfem.GSSmoother(AA)
mfem.PCG(AA, M, B, X, 1, 200, 1e-12, 0.0)
a.RecoverFEMSolution(X, b, x)
-# extract vertices and solution as numpy array
+# Extract vertices and solution as numpy arrays
verts = mesh.GetVertexArray()
sol = x.GetDataArray()
-# plot solution using Matplotlib
-
+# Plot the solution using matplotlib
import matplotlib.pyplot as plt
import matplotlib.tri as tri
triang = tri.Triangulation(verts[:,0], verts[:,1])
-fig1, ax1 = plt.subplots()
-ax1.set_aspect('equal')
-tpc = ax1.tripcolor(triang, sol, shading='gouraud')
-fig1.colorbar(tpc)
+fig, ax = plt.subplots()
+ax.set_aspect('equal')
+tpc = ax.tripcolor(triang, sol, shading='gouraud')
+fig.colorbar(tpc)
plt.show()
```
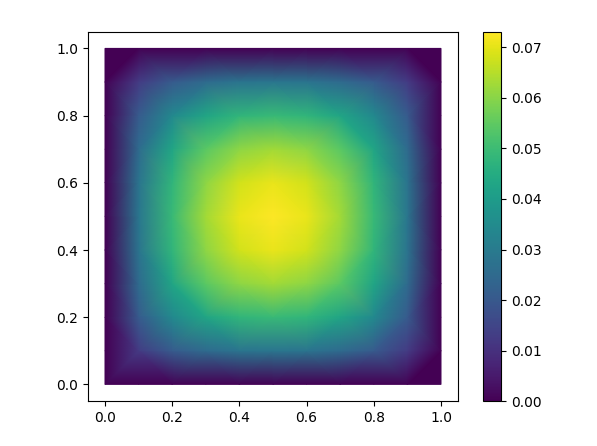
diff --git a/ci_scripts/add_cuda_10_1.sh b/ci_scripts/add_cuda_10_1.sh
deleted file mode 100755
index fbc8da44d..000000000
--- a/ci_scripts/add_cuda_10_1.sh
+++ /dev/null
@@ -1,19 +0,0 @@
-#!/bin/bash
-set -ev
-
-UBUNTU_VERSION=$(lsb_release -sr)
-UBUNTU_VERSION=ubuntu"${UBUNTU_VERSION//.}"
-CUDA=10.1.105-1
-CUDA_SHORT=10.1
-
-INSTALLER=cuda-repo-${UBUNTU_VERSION}_${CUDA}_amd64.deb
-wget http://developer.download.nvidia.com/compute/cuda/repos/${UBUNTU_VERSION}/x86_64/${INSTALLER}
-sudo dpkg -i ${INSTALLER}
-wget https://developer.download.nvidia.com/compute/cuda/repos/${UBUNTU_VERSION}/x86_64/7fa2af80.pub
-sudo apt-key add 7fa2af80.pub
-sudo apt update -qq
-sudo apt install -y cuda-core-${CUDA_SHORT/./-} cuda-cudart-dev-${CUDA_SHORT/./-} cuda-cufft-dev-${CUDA_SHORT/./-}
-sudo apt clean
-
-
-
diff --git a/ci_scripts/add_cuda_11_1.sh b/ci_scripts/add_cuda_11_1.sh
deleted file mode 100755
index c004d6dcd..000000000
--- a/ci_scripts/add_cuda_11_1.sh
+++ /dev/null
@@ -1,11 +0,0 @@
-#!/bin/bash
-set -ev
-
-wget https://developer.download.nvidia.com/compute/cuda/repos/ubuntu2004/x86_64/cuda-ubuntu2004.pin
-sudo mv cuda-ubuntu2004.pin /etc/apt/preferences.d/cuda-repository-pin-600
-wget https://developer.download.nvidia.com/compute/cuda/11.1.1/local_installers/cuda-repo-ubuntu2004-11-1-local_11.1.1-455.32.00-1_amd64.deb
-sudo dpkg -i cuda-repo-ubuntu2004-11-1-local_11.1.1-455.32.00-1_amd64.deb
-sudo apt-key add /var/cuda-repo-ubuntu2004-11-1-local/7fa2af80.pub
-sudo apt-get update
-sudo apt-get -y install cuda
-
diff --git a/ci_scripts/add_cuda_11_5.sh b/ci_scripts/add_cuda_11_5.sh
deleted file mode 100644
index 0c3c23fad..000000000
--- a/ci_scripts/add_cuda_11_5.sh
+++ /dev/null
@@ -1,9 +0,0 @@
-#!/bin/bash
-set -ev
-wget https://developer.download.nvidia.com/compute/cuda/repos/ubuntu2004/x86_64/cuda-ubuntu2004.pin
-sudo mv cuda-ubuntu2004.pin /etc/apt/preferences.d/cuda-repository-pin-600
-wget https://developer.download.nvidia.com/compute/cuda/11.5.0/local_installers/cuda-repo-ubuntu2004-11-5-local_11.5.0-495.29.05-1_amd64.deb
-sudo dpkg -i cuda-repo-ubuntu2004-11-5-local_11.5.0-495.29.05-1_amd64.deb
-sudo apt-key add /var/cuda-repo-ubuntu2004-11-5-local/7fa2af80.pub
-sudo apt-get update
-sudo apt-get -y install cuda
diff --git a/data/compass.geo b/data/compass.geo
new file mode 100644
index 000000000..90572084b
--- /dev/null
+++ b/data/compass.geo
@@ -0,0 +1,118 @@
+SetFactory("OpenCASCADE");
+
+order = 1;
+
+R = 1;
+r = 0.2;
+
+Point(1) = {0,0,0};
+
+Point(2) = {r/Sqrt(2),r/Sqrt(2),0};
+Point(3) = {-r/Sqrt(2),r/Sqrt(2),0};
+Point(4) = {-r/Sqrt(2),-r/Sqrt(2),0};
+Point(5) = {r/Sqrt(2),-r/Sqrt(2),0};
+
+Point(6) = {R,0,0};
+Point(7) = {R/Sqrt(2),R/Sqrt(2),0};
+Point(8) = {0,R,0};
+Point(9) = {-R/Sqrt(2),R/Sqrt(2),0};
+Point(10) = {-R,0,0};
+Point(11) = {-R/Sqrt(2),-R/Sqrt(2),0};
+Point(12) = {0,-R,0};
+Point(13) = {R/Sqrt(2),-R/Sqrt(2),0};
+
+Line(1) = {1,2};
+Line(2) = {1,3};
+Line(3) = {1,4};
+Line(4) = {1,5};
+
+Line(5) = {1,6};
+Line(6) = {1,8};
+Line(7) = {1,10};
+Line(8) = {1,12};
+
+Line(9) = {2,6};
+Line(10) = {2,8};
+Line(11) = {3,8};
+Line(12) = {3,10};
+Line(13) = {4,10};
+Line(14) = {4,12};
+Line(15) = {5,12};
+Line(16) = {5,6};
+
+Line(17) = {6,7};
+Line(18) = {7,8};
+Line(19) = {8,9};
+Line(20) = {9,10};
+Line(21) = {10,11};
+Line(22) = {11,12};
+Line(23) = {12,13};
+Line(24) = {13,6};
+
+Transfinite Curve{1:24} = 2;
+
+Physical Curve("ENE") = {17};
+Physical Curve("NNE") = {18};
+Physical Curve("NNW") = {19};
+Physical Curve("WNW") = {20};
+Physical Curve("WSW") = {21};
+Physical Curve("SSW") = {22};
+Physical Curve("SSE") = {23};
+Physical Curve("ESE") = {24};
+
+Curve Loop(1) = {9,17,18,-10};
+Curve Loop(2) = {11,19,20,-12};
+Curve Loop(3) = {13,21,22,-14};
+Curve Loop(4) = {15,23,24,-16};
+
+Plane Surface(1) = {1};
+Plane Surface(2) = {2};
+Plane Surface(3) = {3};
+Plane Surface(4) = {4};
+
+Transfinite Surface{1} = {2,6,7,8};
+Transfinite Surface{2} = {3,8,9,10};
+Transfinite Surface{3} = {4,10,11,12};
+Transfinite Surface{4} = {5,12,13,6};
+Recombine Surface{1:4};
+
+Physical Surface("Base") = {1,2,3,4};
+
+Curve Loop(5) = {1,10,-6};
+Plane Surface(5) = {5};
+Physical Surface("N Even") = {5};
+
+Curve Loop(6) = {6,-11,-2};
+Plane Surface(6) = {6};
+Physical Surface("N Odd") = {6};
+
+Curve Loop(7) = {2,12,-7};
+Plane Surface(7) = {7};
+Physical Surface("W Even") = {7};
+
+Curve Loop(8) = {7,-13,-3};
+Plane Surface(8) = {8};
+Physical Surface("W Odd") = {8};
+
+Curve Loop(9) = {3,14,-8};
+Plane Surface(9) = {9};
+Physical Surface("S Even") = {9};
+
+Curve Loop(10) = {8,-15,-4};
+Plane Surface(10) = {10};
+Physical Surface("S Odd") = {10};
+
+Curve Loop(11) = {4,16,-5};
+Plane Surface(11) = {11};
+Physical Surface("E Even") = {11};
+
+Curve Loop(12) = {5,-9,-1};
+Plane Surface(12) = {12};
+Physical Surface("E Odd") = {12};
+
+// Generate 2D mesh
+Mesh 2;
+SetOrder order;
+Mesh.MshFileVersion = 2.2;
+
+Save "compass.msh";
diff --git a/data/compass.mesh b/data/compass.mesh
new file mode 100644
index 000000000..8bff51ce5
--- /dev/null
+++ b/data/compass.mesh
@@ -0,0 +1,96 @@
+MFEM mesh v1.3
+
+#
+# MFEM Geometry Types (see mesh/geom.hpp):
+#
+# POINT = 0
+# SEGMENT = 1
+# TRIANGLE = 2
+# SQUARE = 3
+# TETRAHEDRON = 4
+# CUBE = 5
+# PRISM = 6
+#
+
+dimension
+2
+
+elements
+12
+10 2 7 0 1
+11 2 0 7 2
+12 2 9 0 2
+13 2 0 9 3
+14 2 11 0 3
+15 2 0 11 4
+16 2 5 0 4
+17 2 0 5 1
+9 3 1 5 6 7
+9 3 2 7 8 9
+9 3 3 9 10 11
+9 3 4 11 12 5
+
+attribute_sets
+16
+"Base" 1 9
+"E Even" 1 16
+"E Odd" 1 17
+"East" 2 16 17
+"N Even" 1 10
+"N Odd" 1 11
+"North" 2 10 11
+"Rose" 8 10 11 12 13 14 15 16 17
+"Rose Even" 4 10 12 14 16
+"Rose Odd" 4 11 13 15 17
+"S Even" 1 14
+"S Odd" 1 15
+"South" 2 14 15
+"W Even" 1 12
+"W Odd" 1 13
+"West" 2 12 13
+
+boundary
+8
+1 1 5 6
+2 1 6 7
+3 1 7 8
+4 1 8 9
+5 1 9 10
+6 1 10 11
+7 1 11 12
+8 1 12 5
+
+bdr_attribute_sets
+13
+"Boundary" 8 1 2 3 4 5 6 7 8
+"ENE" 1 1
+"ESE" 1 8
+"Eastern Boundary" 2 1 8
+"NNE" 1 2
+"NNW" 1 3
+"Northern Boundary" 2 2 3
+"SSE" 1 7
+"SSW" 1 6
+"Southern Boundary" 2 6 7
+"WNW" 1 4
+"WSW" 1 5
+"Western Boundary" 2 4 5
+
+vertices
+13
+2
+0 0
+0.14142136 0.14142136
+-0.14142136 0.14142136
+-0.14142136 -0.14142136
+0.14142136 -0.14142136
+1 0
+0.70710678 0.70710678
+0 1
+-0.70710678 0.70710678
+-1 0
+-0.70710678 -0.70710678
+0 -1
+0.70710678 -0.70710678
+
+mfem_mesh_end
diff --git a/data/compass.msh b/data/compass.msh
new file mode 100644
index 000000000..f9229490c
--- /dev/null
+++ b/data/compass.msh
@@ -0,0 +1,62 @@
+$MeshFormat
+2.2 0 8
+$EndMeshFormat
+$PhysicalNames
+17
+1 1 "ENE"
+1 2 "NNE"
+1 3 "NNW"
+1 4 "WNW"
+1 5 "WSW"
+1 6 "SSW"
+1 7 "SSE"
+1 8 "ESE"
+2 9 "Base"
+2 10 "N Even"
+2 11 "N Odd"
+2 12 "W Even"
+2 13 "W Odd"
+2 14 "S Even"
+2 15 "S Odd"
+2 16 "E Even"
+2 17 "E Odd"
+$EndPhysicalNames
+$Nodes
+13
+1 0 0 0
+2 0.1414213562373095 0.1414213562373095 0
+3 -0.1414213562373095 0.1414213562373095 0
+4 -0.1414213562373095 -0.1414213562373095 0
+5 0.1414213562373095 -0.1414213562373095 0
+6 1 0 0
+7 0.7071067811865475 0.7071067811865475 0
+8 0 1 0
+9 -0.7071067811865475 0.7071067811865475 0
+10 -1 0 0
+11 -0.7071067811865475 -0.7071067811865475 0
+12 0 -1 0
+13 0.7071067811865475 -0.7071067811865475 0
+$EndNodes
+$Elements
+20
+1 1 2 1 17 6 7
+2 1 2 2 18 7 8
+3 1 2 3 19 8 9
+4 1 2 4 20 9 10
+5 1 2 5 21 10 11
+6 1 2 6 22 11 12
+7 1 2 7 23 12 13
+8 1 2 8 24 13 6
+9 2 2 10 5 1 2 8
+10 2 2 11 6 1 8 3
+11 2 2 12 7 1 3 10
+12 2 2 13 8 1 10 4
+13 2 2 14 9 1 4 12
+14 2 2 15 10 1 12 5
+15 2 2 16 11 1 5 6
+16 2 2 17 12 1 6 2
+17 3 2 9 1 2 6 7 8
+18 3 2 9 2 3 8 9 10
+19 3 2 9 3 4 10 11 12
+20 3 2 9 4 5 12 13 6
+$EndElements
diff --git a/docs/changelog.txt b/docs/changelog.txt
index 07638f9f8..db4f8430f 100644
--- a/docs/changelog.txt
+++ b/docs/changelog.txt
@@ -1,5 +1,22 @@
<<< Change Log. >>>
+2024 08
+ * MFEM 4.7 support
+ - AttributeSets are supported. ex39 and ex39p are added to demonstrate how to use it from Python
+ - Hyperbolic conservation element/face form integrators (hyperbolic.hpp) are supported. ex18.py and
+ ex18.py are updated to conform with the updated C++ examples.
+ - Update SWIG requirement to >= 4.2.1 (required to wrap MFEM routines which use recent C++ features)
+ - Buiding --with-libceed will download libceed=0.12.0, as required by MFEM 4.7
+ - Fixed eltrans::transformback
+ - Improved testing using Github actions
+ - New caller and dispatch yml configulations allows for running a test manually
+ - Test runs automatically for PR and PR update
+ - Test using Python 3.11 is added
+ - Refresh install instruction (Install.md)
+ - Python 3.7 has reached EOL and is no longer supported. This version will support Python 3.8 and above, and
+ will likely be the last version to support Python 3.8.
+
+
2023 11 - 2024 01
* MFEM 4.6 support
- Default MFEM SHA is updated to a version on 11/26/2023 (slightly newer than
diff --git a/docs/install.txt b/docs/install.txt
deleted file mode 100644
index c3a7224f1..000000000
--- a/docs/install.txt
+++ /dev/null
@@ -1,130 +0,0 @@
-# Install Guide
-
-PyMFEM wrapper provides MPI version and non-MPI version of wrapper.
-
-Default pip install installs serial MFEM + wrapper
-
-$ pip install mfem
-or
-$ pip install mfem --no-binary mfem
-
-For other configuration such as parallel version, one can either use --install-option
-flags with pip or download the package as follows and run setup script, manually.
-
-$ pip download mfem --no-binary mfem
-
-In order to see the full list of options, use
-
-$ python setup.py install --help
-
-In below, for the brevity, examples are mostly shown using "python setup.py install" convention.
-When using PIP, each option needs to be passed using --install-option.
-
-## Parallel MFEM
---with-parallel : build both serial and parallel version of MFEM and wrapper
-
-Note: this option turns on building metis and Hypre
-
-## Suitesparse
---with-suitesparse : build MFEM with suitesparse. SuiteSparse needs to be installed separately.
---suitesparse-prefix=
-
-Note: this option turns on building metis in serial
-
-## CUDA
---with-cuda option build MFEM with CUDA. Hypre cuda build is also supported using
---with-cuda-hypre. --cuda-arch can be used to specify cuda compute capablility.
-(See table in https://en.wikipedia.org/wiki/CUDA#Supported_GPUs)
-
--with-cuda : build MFEM using CUDA on
---cuda-arch= : specify cuda compute capability version
---with-cuda-hypre : build Hypre with cuda
-
-(example)
-$ python setup.py install --with-cuda
-$ python setup.py install --with-cuda --with-cuda-hypre
-$ python setup.py install --with-cuda --with-cuda-hypre --cuda-arch=80 (A100)
-$ python setup.py install --with-cuda --with-cuda-hypre --cuda-arch=75 (Turing)
-
-## gslib
---with-gslib : build MFEM with GSlib
-
-Note: this option builds GSlib
-
-## libceed
---with-libceed : build MFEM with libceed
-Note: this option builds libceed
-
-## Specify compilers
---CC c compiler
---CXX c++ compiler
---MPICC mpic compiler
---MPICXX mpic++ compiler
-
-(example)
-Using Intel compiler
-$ python setup.py install --with-parallel --CC=icc, --CXX=icpc, --MPICC=mpiicc, --MPICXX=mpiicpc
-
-## Building MFEM with specific version
-By default, setup.py build MFEM with specific SHA (which is usually the released latest version).
-In order to use the latest MFEM in Github. One can specify the branch name or SHA using mfem-branch
-option.
-
--mfem-branch =
-
-(example)
-$ python setup.py install --mfem-branch=master
-
-## Using MFEM build externally.
-These options are used to link PyMFEM wrapper with existing MFEM library. We need --mfem-source
-and --mfem-prefix
-
---mfem-source : : the location of MFEM source used to build MFEM
---mfem-prefix : : the location of MFEM library. libmfem.so needs to be found in /lib
---mfems-prefix : : (optional) specify serial MFEM location separately
---mfemp-prefix : : (ooptional)specify parallel MFEM location separately
-
-## Blas and Lapack
---with-lapack : build MFEM with lapack
-
- is used for CMAKE call to buid MFEM
---blas_-libraries=
---lapack-libraries=
-
-## Development and testing options
---swig : run swig only
---skip-swig : build without running swig
---skip-ext : skip building external libraries.
---ext-only : build exteranl libraries and exit.
-
-During the development, often we update depenencies (such as MFEM) and edit *.i file.
-
-First clean everything.
-
-$ python setup.py clean --all
-
-Then, build externals alone
-$ python setup.py install --with-parallel --ext-only --mfem-branch="master"
-
-Then, genrate swig wrappers.
-$ python setup.py install --with-parallel --swig --mfem-branch="master"
-
-If you are not happy with the wrapper (*.cxx and *.py), you edit *.i and redo
-the same. When you are happy, build the wrapper. --swig does not clean the
-existing wrapper. So, it will only update wrapper for updated *.i
-
-When building a wrapper, you can use --skip-ext option. By default, it will re-run
-swig to generate entire wrapper codes.
-$ python setup.py install --with-parallel --skip-ext --mfem-branch="master"
-
-If you are sure, you could use --skip-swig option, so that it compiles the wrapper
-codes without re-generating it.
-$ python setup.py install --with-parallel --skip-ext --skip-swig --mfem-branch="master"
-
-
-## Other options
---unverifiedSSL :
- This addresses error relating SSL certificate. Typical error message is
- ""
-
-
diff --git a/examples/ex18.py b/examples/ex18.py
index 772f8e966..1c089ceb5 100644
--- a/examples/ex18.py
+++ b/examples/ex18.py
@@ -3,8 +3,15 @@
This is a version of Example 18 with a simple adaptive mesh
refinement loop.
See c++ version in the MFEM library for more detail
+
+ Sample runs:
+
+ python ex18.py -p 1 -r 2 -o 1 -s 3
+ python ex18.py -p 1 -r 1 -o 3 -s 4
+ python ex18.py -p 1 -r 0 -o 5 -s 6
+ python ex18.py -p 2 -r 1 -o 1 -s 3 -mf
+ python ex18.py -p 2 -r 0 -o 3 -s 3 -mf
'''
-from ex18_common import FE_Evolution, InitialCondition, RiemannSolver, DomainIntegrator, FaceIntegrator
from mfem.common.arg_parser import ArgParser
import mfem.ser as mfem
from mfem.ser import intArray
@@ -13,32 +20,39 @@
from numpy import sqrt, pi, cos, sin, hypot, arctan2
from scipy.special import erfc
-# Equation constant parameters.(using globals to share them with ex18_common)
-import ex18_common
+from ex18_common import (EulerMesh,
+ EulerInitialCondition,
+ DGHyperbolicConservationLaws)
def run(problem=1,
ref_levels=1,
order=3,
ode_solver_type=4,
- t_final=0.5,
+ t_final=2.0,
dt=-0.01,
cfl=0.3,
visualization=True,
vis_steps=50,
+ preassembleWeakDiv=False,
meshfile=''):
- ex18_common.num_equation = 4
- ex18_common.specific_heat_ratio = 1.4
- ex18_common.gas_constant = 1.0
- ex18_common.problem = problem
- num_equation = ex18_common.num_equation
+ specific_heat_ratio = 1.4
+ gas_constant = 1.0
+ IntOrderOffset = 1
# 2. Read the mesh from the given mesh file. This example requires a 2D
# periodic mesh, such as ../data/periodic-square.mesh.
- meshfile = expanduser(join(dirname(__file__), '..', 'data', meshfile))
- mesh = mfem.Mesh(meshfile, 1, 1)
+
+ mesh = EulerMesh(meshfile, problem)
dim = mesh.Dimension()
+ num_equation = dim + 2
+
+ # Refine the mesh to increase the resolution. In this example we do
+ # 'ref_levels' of uniform refinement, where 'ref_levels' is a
+ # command-line parameter.
+ for lev in range(ref_levels):
+ mesh.UniformRefinement()
# 3. Define the ODE solver used for time integration. Several explicit
# Runge-Kutta methods are available.
@@ -48,7 +62,7 @@ def run(problem=1,
elif ode_solver_type == 2:
ode_solver = mfem.RK2Solver(1.0)
elif ode_solver_type == 3:
- ode_solver = mfem.RK3SSolver()
+ ode_solver = mfem.RK3SSPSolver()
elif ode_solver_type == 4:
ode_solver = mfem.RK4Solver()
elif ode_solver_type == 6:
@@ -57,13 +71,7 @@ def run(problem=1,
print("Unknown ODE solver type: " + str(ode_solver_type))
exit
- # 4. Refine the mesh to increase the resolution. In this example we do
- # 'ref_levels' of uniform refinement, where 'ref_levels' is a
- # command-line parameter.
- for lev in range(ref_levels):
- mesh.UniformRefinement()
-
- # 5. Define the discontinuous DG finite element space of the given
+ # 4. Define the discontinuous DG finite element space of the given
# polynomial order on the refined mesh.
fec = mfem.DG_FECollection(order, dim)
@@ -78,70 +86,70 @@ def run(problem=1,
assert fes.GetOrdering() == mfem.Ordering.byNODES, "Ordering must be byNODES"
print("Number of unknowns: " + str(vfes.GetVSize()))
- # 6. Define the initial conditions, save the corresponding mesh and grid
- # functions to a file. This can be opened with GLVis with the -gc option.
- # The solution u has components {density, x-momentum, y-momentum, energy}.
- # These are stored contiguously in the BlockVector u_block.
-
- offsets = [k*vfes.GetNDofs() for k in range(num_equation+1)]
- offsets = mfem.intArray(offsets)
- u_block = mfem.BlockVector(offsets)
- mom = mfem.GridFunction(dfes, u_block, offsets[1])
-
- #
- # Define coefficient using VecotrPyCoefficient and PyCoefficient
- # A user needs to define EvalValue method
- #
- u0 = InitialCondition(num_equation)
- sol = mfem.GridFunction(vfes, u_block.GetData())
+ # 5. Define the initial conditions, save the corresponding mesh and grid
+ # functions to files. These can be opened with GLVis using:
+ # "glvis -m euler-mesh.mesh -g euler-1-init.gf" (for x-momentum).
+ u0 = EulerInitialCondition(problem,
+ specific_heat_ratio,
+ gas_constant)
+ sol = mfem.GridFunction(vfes)
sol.ProjectCoefficient(u0)
- mesh.Print("vortex.mesh", 8)
+ # (Python note): GridFunction pointing to the subset of vector FES.
+ # sol is Vector with dim*fes.GetNDofs()
+ # Since sol.GetDataArray() returns numpy array pointing to the data, we make
+ # Vector from a sub-vector of the returned numpy array and pass it to GridFunction
+ # constructor.
+
+ mom = mfem.GridFunction(dfes, mfem.Vector(
+ sol.GetDataArray()[fes.GetNDofs():]))
+ mesh.Print("euler-mesh.mesh", 8)
+
for k in range(num_equation):
- uk = mfem.GridFunction(fes, u_block.GetBlock(k).GetData())
- sol_name = "vortex-" + str(k) + "-init.gf"
+ uk = mfem.GridFunction(fes, mfem.Vector(
+ sol.GetDataArray()[k*fes.GetNDofs():]))
+ sol_name = "euler-" + str(k) + "-init.gf"
uk.Save(sol_name, 8)
- # 7. Set up the nonlinear form corresponding to the DG discretization of the
- # flux divergence, and assemble the corresponding mass matrix.
- Aflux = mfem.MixedBilinearForm(dfes, fes)
- Aflux.AddDomainIntegrator(DomainIntegrator(dim))
- Aflux.Assemble()
+ # 6. Set up the nonlinear form with euler flux and numerical flux
+ flux = mfem.EulerFlux(dim, specific_heat_ratio)
+ numericalFlux = mfem.RusanovFlux(flux)
+ formIntegrator = mfem.HyperbolicFormIntegrator(
+ numericalFlux, IntOrderOffset)
- A = mfem.NonlinearForm(vfes)
- rsolver = RiemannSolver()
- ii = FaceIntegrator(rsolver, dim)
- A.AddInteriorFaceIntegrator(ii)
-
- # 8. Define the time-dependent evolution operator describing the ODE
- # right-hand side, and perform time-integration (looping over the time
- # iterations, ti, with a time-step dt).
- euler = FE_Evolution(vfes, A, Aflux.SpMat())
+ euler = DGHyperbolicConservationLaws(vfes, formIntegrator,
+ preassembleWeakDivergence=preassembleWeakDiv)
+ # 7. Visualize momentum with its magnitude
if (visualization):
sout = mfem.socketstream("localhost", 19916)
sout.precision(8)
sout << "solution\n" << mesh << mom
+ sout << "window_title 'momentum, t = 0'\n"
+ sout << "view 0 0\n" # view from top
+ sout << "keys jlm\n" # turn off perspective and light, show mesh
sout << "pause\n"
sout.flush()
print("GLVis visualization paused.")
print(" Press space (in the GLVis window) to resume it.")
- # Determine the minimum element size.
- hmin = 0
+ # 8. Time integration
+ hmin = np.inf
if (cfl > 0):
hmin = min([mesh.GetElementSize(i, 1) for i in range(mesh.GetNE())])
+ # Find a safe dt, using a temporary vector. Calling Mult() computes the
+ # maximum char speed at all quadrature points on all faces (and all
+ # elements with -mf).
+ z = mfem.Vector(sol.Size())
+ euler.Mult(sol, z)
+
+ max_char_speed = euler.GetMaxCharSpeed()
+ dt = cfl * hmin / max_char_speed / (2 * order + 1)
+
t = 0.0
euler.SetTime(t)
ode_solver.Init(euler)
- if (cfl > 0):
- # Find a safe dt, using a temporary vector. Calling Mult() computes the
- # maximum char speed at all quadrature points on all faces.
- z = mfem.Vector(A.Width())
- A.Mult(sol, z)
-
- dt = cfl * hmin / ex18_common.max_char_speed / (2*order+1)
# Integrate in time.
done = False
@@ -152,23 +160,29 @@ def run(problem=1,
t, dt_real = ode_solver.Step(sol, t, dt_real)
if (cfl > 0):
- dt = cfl * hmin / ex18_common.max_char_speed / (2*order+1)
+ max_char_speed = euler.GetMaxCharSpeed()
+ dt = cfl * hmin / max_char_speed / (2*order+1)
ti = ti+1
done = (t >= t_final - 1e-8*dt)
if (done or ti % vis_steps == 0):
print("time step: " + str(ti) + ", time: " + "{:g}".format(t))
if (visualization):
- sout << "solution\n" << mesh << mom << flush
-
- # 9. Save the final solution. This output can be viewed later using GLVis:
- # "glvis -m vortex.mesh -g vortex-1-final.gf".
+ sout << "window_title 'momentum, t = " << "{:g}".format(
+ t) << "'\n"
+ sout << "solution\n" << mesh << mom
+ sout.flush()
+
+ # 8. Save the final solution. This output can be viewed later using GLVis:
+ # "glvis -m euler.mesh -g euler-1-final.gf".
+ mesh.Print("euler-mesh-final.mesh", 8)
for k in range(num_equation):
- uk = mfem.GridFunction(fes, u_block.GetBlock(k).GetData())
- sol_name = "vortex-" + str(k) + "-final.gf"
+ uk = mfem.GridFunction(fes, mfem.Vector(
+ sol.GetDataArray()[k*fes.GetNDofs():]))
+ sol_name = "euler-" + str(k) + "-final.gf"
uk.Save(sol_name, 8)
print(" done")
- # 10. Compute the L2 solution error summed for all components.
+ # 9. Compute the L2 solution error summed for all components.
if (t_final == 2.0):
error = sol.ComputeLpError(2., u0)
print("Solution error: " + "{:g}".format(error))
@@ -178,7 +192,7 @@ def run(problem=1,
parser = ArgParser(description='Ex18')
parser.add_argument('-m', '--mesh',
- default='periodic-square.mesh',
+ default='',
action='store', type=str,
help='Mesh file to use.')
parser.add_argument('-p', '--problem',
@@ -203,15 +217,39 @@ def run(problem=1,
parser.add_argument('-c', '--cfl_number',
action='store', default=0.3, type=float,
help="CFL number for timestep calculation.")
- parser.add_argument('-vis', '--visualization',
- action='store_true',
- help='Enable GLVis visualization')
+ parser.add_argument('-novis', '--no_visualization',
+ action='store_true', default=False,
+ help='Disable GLVis visualization')
+ parser.add_argument("-ea", "--element-assembly-divergence",
+ action='store_true', default=False,
+ help="Weak divergence assembly level\n" +
+ " ea - Element assembly with interpolated")
+ parser.add_argument("-mf", "--matrix-free-divergence",
+ action='store_true', default=False,
+ help="Weak divergence assembly level\n" +
+ " mf - Nonlinear assembly in matrix-free manner")
parser.add_argument('-vs', '--visualization-steps',
action='store', default=50, type=float,
help="Visualize every n-th timestep.")
args = parser.parse_args()
+ visualization = not args.no_visualization
+
+ if (not args.matrix_free_divergence and
+ not args.element_assembly_divergence):
+ args.element_assembly_divergence = True
+ args.matrix_free_divergence = False
+ preassembleWeakDiv = True
+
+ elif args.element_assembly_divergence:
+ args.matrix_free_divergence = False
+ preassembleWeakDiv = True
+
+ elif args.matrix_free_divergence:
+ args.element_assembly_divergence = False
+ preassembleWeakDiv = False
+
parser.print_options(args)
run(problem=args.problem,
@@ -221,6 +259,7 @@ def run(problem=1,
t_final=args.t_final,
dt=args.time_step,
cfl=args.cfl_number,
- visualization=args.visualization,
+ visualization=visualization,
vis_steps=args.visualization_steps,
+ preassembleWeakDiv=preassembleWeakDiv,
meshfile=args.mesh)
diff --git a/examples/ex18_common.py b/examples/ex18_common.py
index 9664f718b..5c6fe1ed7 100644
--- a/examples/ex18_common.py
+++ b/examples/ex18_common.py
@@ -3,12 +3,6 @@
This is a python translation of ex18.hpp
- note: following variabls are set from ex18 or ex18p
- problem
- num_equation
- max_char_speed
- specific_heat_ratio;
- gas_constant;
'''
import numpy as np
@@ -19,332 +13,200 @@
else:
import mfem.par as mfem
-num_equation = 0
-specific_heat_ratio = 0
-gas_constant = 0
-problem = 0
-max_char_speed = 0
-
-
-class FE_Evolution(mfem.TimeDependentOperator):
- def __init__(self, vfes, A, A_flux):
- self.dim = vfes.GetFE(0).GetDim()
- self.vfes = vfes
- self.A = A
- self.Aflux = A_flux
- self.Me_inv = mfem.DenseTensor(vfes.GetFE(0).GetDof(),
- vfes.GetFE(0).GetDof(),
- vfes.GetNE())
-
- self.state = mfem.Vector(num_equation)
- self.f = mfem.DenseMatrix(num_equation, self.dim)
- self.flux = mfem.DenseTensor(vfes.GetNDofs(), self.dim, num_equation)
- self.z = mfem.Vector(A.Height())
-
- dof = vfes.GetFE(0).GetDof()
- Me = mfem.DenseMatrix(dof)
- inv = mfem.DenseMatrixInverse(Me)
- mi = mfem.MassIntegrator()
- for i in range(vfes.GetNE()):
- mi.AssembleElementMatrix(vfes.GetFE(
- i), vfes.GetElementTransformation(i), Me)
- inv.Factor()
- inv.GetInverseMatrix(self.Me_inv(i))
- super(FE_Evolution, self).__init__(A.Height())
-
- def GetFlux(self, x, flux):
- state = self.state
- dof = self.flux.SizeI()
- dim = self.flux.SizeJ()
-
- flux_data = []
- for i in range(dof):
- for k in range(num_equation):
- self.state[k] = x[i, k]
- ComputeFlux(state, dim, self.f)
-
- flux_data.append(self.f.GetDataArray().transpose().copy())
- # flux[i].Print()
- # print(self.f.GetDataArray())
- # for d in range(dim):
- # for k in range(num_equation):
- # flux[i, d, k] = self.f[k, d]
-
- mcs = ComputeMaxCharSpeed(state, dim)
- if (mcs > globals()['max_char_speed']):
- globals()['max_char_speed'] = mcs
-
- flux.Assign(np.stack(flux_data))
- #print("max char speed", globals()['max_char_speed'])
+from os.path import expanduser, join, dirname
- def Mult(self, x, y):
- globals()['max_char_speed'] = 0.
- num_equation = globals()['num_equation']
- # 1. Create the vector z with the face terms -.
- self.A.Mult(x, self.z)
-
- # 2. Add the element terms.
- # i. computing the flux approximately as a grid function by interpolating
- # at the solution nodes.
- # ii. multiplying this grid function by a (constant) mixed bilinear form for
- # each of the num_equation, computing (F(u), grad(w)) for each equation.
-
- xmat = mfem.DenseMatrix(
- x.GetData(), self.vfes.GetNDofs(), num_equation)
- self.GetFlux(xmat, self.flux)
-
- for k in range(num_equation):
- fk = mfem.Vector(self.flux[k].GetData(),
- self.dim * self.vfes.GetNDofs())
- o = k * self.vfes.GetNDofs()
- zk = self.z[o: o+self.vfes.GetNDofs()]
- self.Aflux.AddMult(fk, zk)
-
- # 3. Multiply element-wise by the inverse mass matrices.
- zval = mfem.Vector()
- vdofs = mfem.intArray()
- dof = self.vfes.GetFE(0).GetDof()
- zmat = mfem.DenseMatrix()
- ymat = mfem.DenseMatrix(dof, num_equation)
- for i in range(self.vfes.GetNE()):
- # Return the vdofs ordered byNODES
- vdofs = mfem.intArray(self.vfes.GetElementVDofs(i))
- self.z.GetSubVector(vdofs, zval)
- zmat.UseExternalData(zval.GetData(), dof, num_equation)
- mfem.Mult(self.Me_inv[i], zmat, ymat)
- y.SetSubVector(vdofs, ymat.GetData())
-
-
-class DomainIntegrator(mfem.PyBilinearFormIntegrator):
- def __init__(self, dim):
- num_equation = globals()['num_equation']
- self.flux = mfem.DenseMatrix(num_equation, dim)
- self.shape = mfem.Vector()
- self.dshapedr = mfem.DenseMatrix()
- self.dshapedx = mfem.DenseMatrix()
- super(DomainIntegrator, self).__init__()
-
- def AssembleElementMatrix2(self, trial_fe, test_fe, Tr, elmat):
- # Assemble the form (vec(v), grad(w))
-
- # Trial space = vector L2 space (mesh dim)
- # Test space = scalar L2 space
-
- dof_trial = trial_fe.GetDof()
- dof_test = test_fe.GetDof()
- dim = trial_fe.GetDim()
-
- self.shape.SetSize(dof_trial)
- self.dshapedr.SetSize(dof_test, dim)
- self.dshapedx.SetSize(dof_test, dim)
-
- elmat.SetSize(dof_test, dof_trial * dim)
- elmat.Assign(0.0)
-
- maxorder = max(trial_fe.GetOrder(), test_fe.GetOrder())
- intorder = 2 * maxorder
- ir = mfem.IntRules.Get(trial_fe.GetGeomType(), intorder)
-
- for i in range(ir.GetNPoints()):
- ip = ir.IntPoint(i)
-
- # Calculate the shape functions
- trial_fe.CalcShape(ip, self.shape)
- self.shape *= ip.weight
-
- # Compute the physical gradients of the test functions
- Tr.SetIntPoint(ip)
- test_fe.CalcDShape(ip, self.dshapedr)
- mfem.Mult(self.dshapedr, Tr.AdjugateJacobian(), self.dshapedx)
-
- for d in range(dim):
- for j in range(dof_test):
- for k in range(dof_trial):
- elmat[j, k + d * dof_trial] += self.shape[k] * \
- self.dshapedx[j, d]
-
-
-class FaceIntegrator(mfem.PyNonlinearFormIntegrator):
- def __init__(self, rsolver, dim):
- self.rsolver = rsolver
- self.shape1 = mfem.Vector()
- self.shape2 = mfem.Vector()
- self.funval1 = mfem.Vector(num_equation)
- self.funval2 = mfem.Vector(num_equation)
- self.nor = mfem.Vector(dim)
- self.fluxN = mfem.Vector(num_equation)
- self.eip1 = mfem.IntegrationPoint()
- self.eip2 = mfem.IntegrationPoint()
- super(FaceIntegrator, self).__init__()
-
- self.fluxNA = np.atleast_2d(self.fluxN.GetDataArray())
-
- def AssembleFaceVector(self, el1, el2, Tr, elfun, elvect):
- num_equation = globals()['num_equation']
- # Compute the term on the interior faces.
- dof1 = el1.GetDof()
- dof2 = el2.GetDof()
-
- self.shape1.SetSize(dof1)
- self.shape2.SetSize(dof2)
-
- elvect.SetSize((dof1 + dof2) * num_equation)
- elvect.Assign(0.0)
-
- elfun1_mat = mfem.DenseMatrix(elfun.GetData(), dof1, num_equation)
- elfun2_mat = mfem.DenseMatrix(
- elfun[dof1*num_equation:].GetData(), dof2, num_equation)
-
- elvect1_mat = mfem.DenseMatrix(elvect.GetData(), dof1, num_equation)
- elvect2_mat = mfem.DenseMatrix(
- elvect[dof1*num_equation:].GetData(), dof2, num_equation)
-
- # Integration order calculation from DGTraceIntegrator
- if (Tr.Elem2No >= 0):
- intorder = (min(Tr.Elem1.OrderW(), Tr.Elem2.OrderW()) +
- 2*max(el1.GetOrder(), el2.GetOrder()))
+class DGHyperbolicConservationLaws(mfem.TimeDependentOperator):
+ def __init__(self, vfes_, formIntegrator_, preassembleWeakDivergence=True):
+
+ super(DGHyperbolicConservationLaws, self).__init__(
+ vfes_.GetTrueVSize())
+ self.num_equations = formIntegrator_.num_equations
+ self.vfes = vfes_
+ self.dim = vfes_.GetMesh().SpaceDimension()
+ self.formIntegrator = formIntegrator_
+
+ self.z = mfem.Vector(vfes_.GetTrueVSize())
+
+ self.weakdiv = None
+ self.max_char_speed = None
+
+ self.ComputeInvMass()
+
+ if mfem_mode == 'serial':
+ self.nonlinearForm = mfem.NonlinearForm(self.vfes)
+ else:
+ if isinstance(self.vfes, mfem.ParFiniteElementSpace):
+ self.nonlinearForm = mfem.ParNonlinearForm(self.vfes)
+ else:
+ self.nonlinearForm = mfem.NonlinearForm(self.vfes)
+
+ if preassembleWeakDivergence:
+ self.ComputeWeakDivergence()
else:
- intorder = Tr.Elem1.OrderW() + 2*el1.GetOrder()
+ self.nonlinearForm.AddDomainIntegrator(self.formIntegrator)
+
+ self.nonlinearForm.AddInteriorFaceIntegrator(self.formIntegrator)
+ self.nonlinearForm.UseExternalIntegrators()
+
+ def GetMaxCharSpeed(self):
+ return self.max_char_speed
- if (el1.Space() == mfem.FunctionSpace().Pk):
- intorder += 1
+ def ComputeInvMass(self):
+ inv_mass = mfem.InverseIntegrator(mfem.MassIntegrator())
- ir = mfem.IntRules.Get(Tr.GetGeometryType(), int(intorder))
+ self.invmass = [None]*self.vfes.GetNE()
+ for i in range(self.vfes.GetNE()):
+ dof = self.vfes.GetFE(i).GetDof()
+ self.invmass[i] = mfem.DenseMatrix(dof)
+ inv_mass.AssembleElementMatrix(self.vfes.GetFE(i),
+ self.vfes.GetElementTransformation(
+ i),
+ self.invmass[i])
+
+ def ComputeWeakDivergence(self):
+ weak_div = mfem.TransposeIntegrator(mfem.GradientIntegrator())
+
+ weakdiv_bynodes = mfem.DenseMatrix()
- mat1A = elvect1_mat.GetDataArray()
- mat2A = elvect2_mat.GetDataArray()
- shape1A = np.atleast_2d(self.shape1.GetDataArray())
- shape2A = np.atleast_2d(self.shape2.GetDataArray())
+ self.weakdiv = [None]*self.vfes.GetNE()
- for i in range(ir.GetNPoints()):
- ip = ir.IntPoint(i)
- Tr.Loc1.Transform(ip, self.eip1)
- Tr.Loc2.Transform(ip, self.eip2)
+ for i in range(self.vfes.GetNE()):
+ dof = self.vfes.GetFE(i).GetDof()
+ weakdiv_bynodes.SetSize(dof, dof*self.dim)
+ weak_div.AssembleElementMatrix2(self.vfes.GetFE(i),
+ self.vfes.GetFE(i),
+ self.vfes.GetElementTransformation(
+ i),
+ weakdiv_bynodes)
+ self.weakdiv[i] = mfem.DenseMatrix()
+ self.weakdiv[i].SetSize(dof, dof*self.dim)
+
+ # Reorder so that trial space is ByDim.
+ # This makes applying weak divergence to flux value simpler.
+ for j in range(dof):
+ for d in range(self.dim):
+ self.weakdiv[i].SetCol(
+ j*self.dim + d, weakdiv_bynodes.GetColumn(d*dof + j))
- # Calculate basis functions on both elements at the face
- el1.CalcShape(self.eip1, self.shape1)
- el2.CalcShape(self.eip2, self.shape2)
+ def Mult(self, x, y):
+ # 0. Reset wavespeed computation before operator application.
+ self.formIntegrator.ResetMaxCharSpeed()
+
+ # 1. Apply Nonlinear form to obtain an auxiliary result
+ # z = - _e
+ # If weak-divergence is not preassembled, we also have weak-divergence
+ # z = - _e + (F(u_h), ∇v)
+ self.nonlinearForm.Mult(x, self.z)
+ #print("!!!!", self.weakdiv)
+ if self.weakdiv is not None: # if weak divergence is pre-assembled
+ # Apply weak divergence to F(u_h), and inverse mass to z_loc + weakdiv_loc
+
+ current_state = mfem.Vector() # view of current state at a node
+ current_flux = mfem.DenseMatrix() # flux of current state
+
+ # element flux value. Whose column is ordered by dim.
+ flux = mfem.DenseMatrix()
+ # view of current states in an element, dof x num_eq
+ current_xmat = mfem.DenseMatrix()
+ # view of element auxiliary result, dof x num_eq
+ current_zmat = mfem.DenseMatrix()
+ current_ymat = mfem.DenseMatrix() # view of element result, dof x num_eq
+
+ fluxFunction = self.formIntegrator.GetFluxFunction()
+
+ xval = mfem.Vector()
+ zval = mfem.Vector()
+ flux_vec = mfem.Vector()
+
+ for i in range(self.vfes.GetNE()):
+ Tr = self.vfes.GetElementTransformation(i)
+ dof = self.vfes.GetFE(i).GetDof()
+ vdofs = mfem.intArray(self.vfes.GetElementVDofs(i))
+
+ x.GetSubVector(vdofs, xval)
+ current_xmat.UseExternalData(
+ xval.GetData(), dof, self.num_equations)
+
+ #
+ # Python Note:
+ # C++ code access to array data with offset is done bu GetData() + offset
+ # In Python, the same can be done by using numpy array generated from Vector::GetDataArray(),
+ #
+ # array = vec.GetDataArray()
+ # new_data_pointer = mfem.Vector(array[10:]).GetData()
+ #
+ # note that the above does not work if mfem.Vector is replaced by mfem.DenseMatrix
+ # This is because, while MFEM stores data in colume-major, Python numpy store raw-major.
+ #
+
+ flux.SetSize(self.num_equations, self.dim*dof)
+ flux_vec = mfem.Vector(
+ flux.GetData(), self.num_equations*self.dim*dof)
+ data = flux_vec.GetDataArray()
+
+ for j in range(dof): # compute flux for all nodes in the element
+ current_xmat.GetRow(j, current_state)
+
+ data_ptr = mfem.Vector(
+ data[self.num_equations*self.dim*j:]).GetData()
+ current_flux = mfem.DenseMatrix(data_ptr,
+ self.num_equations, dof)
+ fluxFunction.ComputeFlux(current_state, Tr, current_flux)
+
+ # Compute weak-divergence and add it to auxiliary result, z
+ # Recalling that weakdiv is reordered by dim, we can apply
+ # weak-divergence to the transpose of flux.
+ self.z.GetSubVector(vdofs, zval)
+ current_zmat.UseExternalData(
+ zval.GetData(), dof, self.num_equations)
+ mfem.AddMult_a_ABt(1.0, self.weakdiv[i], flux, current_zmat)
+
+ # Apply inverse mass to auxiliary result to obtain the final result
+ current_ymat.SetSize(dof, self.num_equations)
+ mfem.Mult(self.invmass[i], current_zmat, current_ymat)
+ y.SetSubVector(vdofs, current_ymat.GetData())
- # Interpolate elfun at the point
- elfun1_mat.MultTranspose(self.shape1, self.funval1)
- elfun2_mat.MultTranspose(self.shape2, self.funval2)
- Tr.Face.SetIntPoint(ip)
-
- # Get the normal vector and the flux on the face
-
- mfem.CalcOrtho(Tr.Face.Jacobian(), self.nor)
-
- mcs = self.rsolver.Eval(
- self.funval1, self.funval2, self.nor, self.fluxN)
-
- # Update max char speed
- if mcs > globals()['max_char_speed']:
- globals()['max_char_speed'] = mcs
-
- self.fluxN *= ip.weight
-
- #
- mat1A -= shape1A.transpose().dot(self.fluxNA)
- mat2A += shape2A.transpose().dot(self.fluxNA)
- '''
- for k in range(num_equation):
- for s in range(dof1):
- elvect1_mat[s, k] -= self.fluxN[k] * self.shape1[s]
- for s in range(dof2):
- elvect2_mat[s, k] += self.fluxN[k] * self.shape2[s]
- '''
-
-
-class RiemannSolver(object):
- def __init__(self):
- num_equation = globals()['num_equation']
- self.flux1 = mfem.Vector(num_equation)
- self.flux2 = mfem.Vector(num_equation)
-
- def Eval(self, state1, state2, nor, flux):
-
- # NOTE: nor in general is not a unit normal
- dim = nor.Size()
-
- assert StateIsPhysical(state1, dim), ""
- assert StateIsPhysical(state2, dim), ""
-
- maxE1 = ComputeMaxCharSpeed(state1, dim)
- maxE2 = ComputeMaxCharSpeed(state2, dim)
- maxE = max(maxE1, maxE2)
-
- ComputeFluxDotN(state1, nor, self.flux1)
- ComputeFluxDotN(state2, nor, self.flux2)
-
- #normag = np.sqrt(np.sum(nor.GetDataArray()**2))
- normag = nor.Norml2()
-
- '''
- for i in range(num_equation):
- flux[i] = (0.5 * (self.flux1[i] + self.flux2[i])
- - 0.5 * maxE * (state2[i] - state1[i]) * normag)
- '''
- f = (0.5 * (self.flux1.GetDataArray() + self.flux2.GetDataArray())
- - 0.5 * maxE * (state2.GetDataArray() - state1.GetDataArray()) * normag)
- flux.Assign(f)
-
- return maxE
-
-
-def StateIsPhysical(state, dim):
- specific_heat_ratio = globals()["specific_heat_ratio"]
-
- den = state[0]
- #den_vel = state.GetDataArray()[1:1+dim]
- den_energy = state[1 + dim]
-
- if (den < 0):
- print("Negative density: " + str(state.GetDataArray()))
- return False
- if (den_energy <= 0):
- print("Negative energy: " + str(state.GetDataArray()))
- return False
-
- #den_vel2 = np.sum(den_vel**2)/den
- den_vel2 = (state[1:1+dim].Norml2())**2/den
- pres = (specific_heat_ratio - 1.0) * (den_energy - 0.5 * den_vel2)
- if (pres <= 0):
- print("Negative pressure: " + str(state.GetDataArray()))
- return False
- return True
-
-
-class InitialCondition(mfem.VectorPyCoefficient):
- def __init__(self, dim):
- mfem.VectorPyCoefficient.__init__(self, dim)
-
- def EvalValue(self, x):
- dim = x.shape[0]
- assert dim == 2, ""
- problem = globals()['problem']
- if (problem == 1):
- # "Fast vortex"
- radius = 0.2
- Minf = 0.5
- beta = 1. / 5.
- elif (problem == 2):
- # "Slow vortex"
- radius = 0.2
- Minf = 0.05
- beta = 1. / 50.
else:
- assert False, "Cannot recognize problem. Options are: 1 - fast vortex, 2 - slow vortex"
+ # Apply block inverse mass
+ zval = mfem.Vector() # / z_loc, dof*num_eq
+
+ # view of element auxiliary result, dof x num_eq
+ current_zmat = mfem.DenseMatrix()
+ current_ymat = mfem.DenseMatrix() # view of element result, dof x num_eq
+
+ for i in range(self.vfes.GetNE()):
+ dof = self.vfes.GetFE(i).GetDof()
+ vdofs = mfem.intArray(self.vfes.GetElementVDofs(i))
+ self.z.GetSubVector(vdofs, zval)
+ current_zmat.UseExternalData(
+ zval.GetData(), dof, self.num_equations)
+ current_ymat.SetSize(dof, self.num_equations)
+ mfem.Mult(self.invmass[i], current_zmat, current_ymat)
+ y.SetSubVector(vdofs, current_ymat.GetData())
+
+ self.max_char_speed = self.formIntegrator.GetMaxCharSpeed()
+
+ def Update(self):
+ self.nonlinearForm.Update()
+ height = self.nonlinearForm.Height()
+ width = height
+ self.z.SetSize(height)
+
+ ComputeInvMass()
+ if self.weakdiv is None:
+ self.ComputeWeakDivergence()
+
+
+def GetMovingVortexInit(radius, Minf, beta, gas_constant, specific_heat_ratio):
+ def func(x, y):
xc = 0.0
yc = 0.0
# Nice units
vel_inf = 1.
den_inf = 1.
- specific_heat_ratio = globals()["specific_heat_ratio"]
- gas_constant = globals()["gas_constant"]
-
pres_inf = (den_inf / specific_heat_ratio) * \
(vel_inf / Minf) * (vel_inf / Minf)
temp_inf = pres_inf / (den_inf * gas_constant)
@@ -370,74 +232,80 @@ def EvalValue(self, x):
pres = den * gas_constant * temp
energy = shrinv1 * pres / den + 0.5 * vel2
- y = np.array([den, den * velX, den * velY, den * energy])
- return y
-
-
-def ComputePressure(state, dim):
- den = state[0]
- #den_vel = state.GetDataArray()[1:1+dim]
- den_energy = state[1 + dim]
-
- specific_heat_ratio = globals()["specific_heat_ratio"]
- #den_vel2 = np.sum(den_vel**2)/den
- den_vel2 = (state[1:1+dim].Norml2())**2/den
- pres = (specific_heat_ratio - 1.0) * (den_energy - 0.5 * den_vel2)
-
- return pres
-
-
-def ComputeFlux(state, dim, flux):
- den = state[0]
- den_vel = state.GetDataArray()[1:1+dim]
- den_energy = state[1 + dim]
-
- assert StateIsPhysical(state, dim), ""
-
- pres = ComputePressure(state, dim)
-
- den_vel2 = np.atleast_2d(den_vel)
- fluxA = flux.GetDataArray()
- fluxA[0, :] = den_vel
- fluxA[1:1+dim, :] = den_vel2.transpose().dot(den_vel2) / den
- for d in range(dim):
- fluxA[1+d, d] += pres
+ y[0] = den
+ y[1] = den * velX
+ y[2] = den * velY
+ y[3] = den * energy
- H = (den_energy + pres) / den
- flux.GetDataArray()[1+dim, :] = den_vel * H
+ return func
-def ComputeFluxDotN(state, nor, fluxN):
- # NOTE: nor in general is not a unit normal
- dim = nor.Size()
- nor = nor.GetDataArray()
- fluxN = fluxN.GetDataArray()
+def EulerMesh(meshfile, problem):
+ if meshfile == '':
+ if problem in (1, 2, 3):
+ meshfile = "periodic-square.mesh"
- den = state[0]
- den_vel = state.GetDataArray()[1:1+dim]
- den_energy = state[1 + dim]
+ elif problem == 4:
+ meshfile = "periodic-segment.mesh"
- assert StateIsPhysical(state, dim), ""
-
- pres = ComputePressure(state, dim)
-
- den_velN = den_vel.dot(nor)
-
- fluxN[0] = den_velN
- fluxN[1:1+dim] = den_velN * den_vel / den + pres * nor
-
- H = (den_energy + pres) / den
- fluxN[1+dim] = den_velN * H
-
-
-def ComputeMaxCharSpeed(state, dim):
- specific_heat_ratio = globals()["specific_heat_ratio"]
-
- den = state[0]
- den_vel2 = (state[1:1+dim].Norml2())**2/den
- pres = ComputePressure(state, dim)
-
- sound = np.sqrt(specific_heat_ratio * pres / den)
- vel = np.sqrt(den_vel2 / den)
-
- return vel + sound
+ else:
+ assert False, "Default mesh file not given for problem = " + \
+ str(problem)
+
+ meshfile = expanduser(join(dirname(__file__), '..', 'data', meshfile))
+
+ return mfem.Mesh(meshfile, 1, 1)
+
+# Initial condition
+
+
+def EulerInitialCondition(problem, specific_heat_ratio, gas_constant):
+
+ if problem == 1:
+ # fast moving vortex
+ func = GetMovingVortexInit(0.2, 0.5, 1. / 5., gas_constant,
+ specific_heat_ratio)
+ return mfem.jit.vector(vdim=4, interface="c++")(func)
+
+ elif problem == 2:
+ # slow moving vortex
+ func = GetMovingVortexInit(0.2, 0.05, 1. / 50., gas_constant,
+ specific_heat_ratio)
+ return mfem.jit.vector(vdim=4, interface="c++")(func)
+
+ elif problem == 3:
+ # moving sine wave
+ @ mfem.jit.vector(vdim=4, interface="c++")
+ def func(x, y):
+ assert len(x) > 2, "2D is not supportd for this probl"
+ density = 1.0 + 0.2 * np.sin(np.pi*(x[0]+x[1]))
+ velocity_x = 0.7
+ velocity_y = 0.3
+ pressure = 1.0
+ energy = (pressure / (1.4 - 1.0) +
+ density * 0.5 * (velocity_x * velocity_x + velocity_y * velocity_y))
+
+ y[0] = density
+ y[1] = density * velocity_x
+ y[2] = density * velocity_y
+ y[3] = energy
+
+ return func
+
+ elif problem == 4:
+ @ mfem.jit.vector(vdim=3, interface="c++")
+ def func(x, y):
+ density = 1.0 + 0.2 * np.sin(np.pi * 2 * x[0])
+ velocity_x = 1.0
+ pressure = 1.0
+ energy = pressure / (1.4 - 1.0) + density * \
+ 0.5 * (velocity_x * velocity_x)
+
+ y[0] = density
+ y[1] = density * velocity_x
+ y[2] = energy
+
+ return func
+
+ else:
+ assert False, "Problem Undefined"
diff --git a/examples/ex18p.py b/examples/ex18p.py
index ac57fe237..3253af540 100644
--- a/examples/ex18p.py
+++ b/examples/ex18p.py
@@ -3,10 +3,17 @@
This is a version of Example 18 with a simple adaptive mesh
refinement loop.
See c++ version in the MFEM library for more detail
+ Sample runs:
+
+ mpirun -np 4 python ex18p.py -p 1 -rs 2 -rp 1 -o 1 -s 3
+ mpirun -np 4 python ex18p.py -p 1 -rs 1 -rp 1 -o 3 -s 4
+ mpirun -np 4 python ex18p.py -p 1 -rs 1 -rp 1 -o 5 -s 6
+ mpirun -np 4 python ex18p.py -p 2 -rs 1 -rp 1 -o 1 -s 3 -mf
+ mpirun -np 4 python ex18p.py -p 2 -rs 1 -rp 1 -o 3 -s 3 -mf
+
'''
import mfem.par as mfem
-from ex18_common import FE_Evolution, InitialCondition, RiemannSolver, DomainIntegrator, FaceIntegrator
from mfem.common.arg_parser import ArgParser
from os.path import expanduser, join, dirname
@@ -14,8 +21,9 @@
from numpy import sqrt, pi, cos, sin, hypot, arctan2
from scipy.special import erfc
-# Equation constant parameters.(using globals to share them with ex18_common)
-import ex18_common
+from ex18_common import (EulerMesh,
+ EulerInitialCondition,
+ DGHyperbolicConservationLaws)
# 1. Initialize MPI.from mpi4py import MPI
@@ -23,231 +31,287 @@
num_procs = MPI.COMM_WORLD.size
myid = MPI.COMM_WORLD.rank
-parser = ArgParser(description='Ex18p')
-parser.add_argument('-m', '--mesh',
- default='periodic-square.mesh',
- action='store', type=str,
- help='Mesh file to use.')
-parser.add_argument('-p', '--problem',
- action='store', default=1, type=int,
- help='Problem setup to use. See options in velocity_function().')
-parser.add_argument('-rs', '--refine_serial',
- action='store', default=0, type=int,
- help="Number of times to refine the mesh uniformly before parallel.")
-parser.add_argument('-rp', '--refine_parallel',
- action='store', default=1, type=int,
- help="Number of times to refine the mesh uniformly after parallel.")
-parser.add_argument('-o', '--order',
- action='store', default=3, type=int,
- help="Finite element order (polynomial degree)")
-parser.add_argument('-s', '--ode_solver',
- action='store', default=4, type=int,
- help="ODE solver: 1 - Forward Euler,\n\t" +
- " 2 - RK2 SSP, 3 - RK3 SSP, 4 - RK4, 6 - RK6.")
-parser.add_argument('-tf', '--t_final',
- action='store', default=2.0, type=float,
- help="Final time; start time is 0.")
-parser.add_argument("-dt", "--time_step",
- action='store', default=-0.01, type=float,
- help="Time step.")
-parser.add_argument('-c', '--cfl_number',
- action='store', default=0.3, type=float,
- help="CFL number for timestep calculation.")
-parser.add_argument('-vis', '--visualization',
- action='store_true',
- help='Enable GLVis visualization')
-parser.add_argument('-vs', '--visualization-steps',
- action='store', default=50, type=float,
- help="Visualize every n-th timestep.")
-
-args = parser.parse_args()
-mesh = args.mesh
-ser_ref_levels = args.refine_serial
-par_ref_levels = args.refine_parallel
-order = args.order
-ode_solver_type = args.ode_solver
-t_final = args.t_final
-dt = args.time_step
-cfl = args.cfl_number
-visualization = args.visualization
-vis_steps = args.visualization_steps
-
-if myid == 0:
- parser.print_options(args)
-
-device = mfem.Device('cpu')
-if myid == 0:
- device.Print()
-
-ex18_common.num_equation = 4
-ex18_common.specific_heat_ratio = 1.4
-ex18_common.gas_constant = 1.0
-ex18_common.problem = args.problem
-num_equation = ex18_common.num_equation
-
-
-# 3. Read the mesh from the given mesh file. This example requires a 2D
-# periodic mesh, such as ../data/periodic-square.mesh.
-meshfile = expanduser(join(dirname(__file__), '..', 'data', mesh))
-mesh = mfem.Mesh(meshfile, 1, 1)
-dim = mesh.Dimension()
-
-# 4. Define the ODE solver used for time integration. Several explicit
-# Runge-Kutta methods are available.
-ode_solver = None
-if ode_solver_type == 1:
- ode_solver = mfem.ForwardEulerSolver()
-elif ode_solver_type == 2:
- ode_solver = mfem.RK2Solver(1.0)
-elif ode_solver_type == 3:
- ode_solver = mfem.RK3SSolver()
-elif ode_solver_type == 4:
- ode_solver = mfem.RK4Solver()
-elif ode_solver_type == 6:
- ode_solver = mfem.RK6Solver()
-else:
- print("Unknown ODE solver type: " + str(ode_solver_type))
- exit
-
-# 5. Refine the mesh in serial to increase the resolution. In this example
-# we do 'ser_ref_levels' of uniform refinement, where 'ser_ref_levels' is
-# a command-line parameter.
-for lev in range(ser_ref_levels):
- mesh.UniformRefinement()
-
-# 6. Define a parallel mesh by a partitioning of the serial mesh. Refine
-# this mesh further in parallel to increase the resolution. Once the
-# parallel mesh is defined, the serial mesh can be deleted.
-
-pmesh = mfem.ParMesh(MPI.COMM_WORLD, mesh)
-del mesh
-for lev in range(par_ref_levels):
- pmesh.UniformRefinement()
-
-# 7. Define the discontinuous DG finite element space of the given
-# polynomial order on the refined mesh.
-fec = mfem.DG_FECollection(order, dim)
-# Finite element space for a scalar (thermodynamic quantity)
-fes = mfem.ParFiniteElementSpace(pmesh, fec)
-# Finite element space for a mesh-dim vector quantity (momentum)
-dfes = mfem.ParFiniteElementSpace(pmesh, fec, dim, mfem.Ordering.byNODES)
-# Finite element space for all variables together (total thermodynamic state)
-vfes = mfem.ParFiniteElementSpace(
- pmesh, fec, num_equation, mfem.Ordering.byNODES)
-
-assert fes.GetOrdering() == mfem.Ordering.byNODES, "Ordering must be byNODES"
-glob_size = vfes.GlobalTrueVSize()
-if myid == 0:
- print("Number of unknowns: " + str(glob_size))
-
-# 8. Define the initial conditions, save the corresponding mesh and grid
-# functions to a file. This can be opened with GLVis with the -gc option.
-# The solution u has components {density, x-momentum, y-momentum, energy}.
-# These are stored contiguously in the BlockVector u_block.
-
-offsets = [k*vfes.GetNDofs() for k in range(num_equation+1)]
-offsets = mfem.intArray(offsets)
-u_block = mfem.BlockVector(offsets)
-
-# Momentum grid function on dfes for visualization.
-mom = mfem.ParGridFunction(dfes, u_block, offsets[1])
-
-# Initialize the state.
-u0 = InitialCondition(num_equation)
-sol = mfem.ParGridFunction(vfes, u_block.GetData())
-sol.ProjectCoefficient(u0)
-
-smyid = '{:0>6d}'.format(myid)
-pmesh.Print("vortex-mesh."+smyid, 8)
-for k in range(num_equation):
- uk = mfem.ParGridFunction(fes, u_block.GetBlock(k).GetData())
- sol_name = "vortex-" + str(k) + "-init."+smyid
- uk.Save(sol_name, 8)
-
-# 9. Set up the nonlinear form corresponding to the DG discretization of the
-# flux divergence, and assemble the corresponding mass matrix.
-Aflux = mfem.MixedBilinearForm(dfes, fes)
-Aflux.AddDomainIntegrator(DomainIntegrator(dim))
-Aflux.Assemble()
-
-A = mfem.ParNonlinearForm(vfes)
-rsolver = RiemannSolver()
-ii = FaceIntegrator(rsolver, dim)
-A.AddInteriorFaceIntegrator(ii)
-
-# 10. Define the time-dependent evolution operator describing the ODE
-# right-hand side, and perform time-integration (looping over the time
-# iterations, ti, with a time-step dt).
-euler = FE_Evolution(vfes, A, Aflux.SpMat())
-
-if (visualization):
- MPI.COMM_WORLD.Barrier()
- sout = mfem.socketstream("localhost", 19916)
- sout.send_text("parallel " + str(num_procs) + " " + str(myid))
- sout.precision(8)
- sout.send_solution(pmesh, mom)
- sout.send_text("pause")
- sout.flush()
+
+def run(problem=1,
+ ser_ref_levels=0,
+ par_ref_levels=1,
+ order=3,
+ ode_solver_type=4,
+ t_final=2.0,
+ dt=-0.01,
+ cfl=0.3,
+ visualization=True,
+ vis_steps=50,
+ preassembleWeakDiv=False,
+ meshfile=''):
+
+ specific_heat_ratio = 1.4
+ gas_constant = 1.0
+ IntOrderOffset = 1
+
+ device = mfem.Device('cpu')
if myid == 0:
- print("GLVis visualization paused.")
- print(" Press space (in the GLVis window) to resume it.")
-
-# Determine the minimum element size.
-my_hmin = 0
-if (cfl > 0):
- my_hmin = min([pmesh.GetElementSize(i, 1) for i in range(pmesh.GetNE())])
-hmin = MPI.COMM_WORLD.allreduce(my_hmin, op=MPI.MIN)
-
-t = 0.0
-euler.SetTime(t)
-ode_solver.Init(euler)
-if (cfl > 0):
- # Find a safe dt, using a temporary vector. Calling Mult() computes the
- # maximum char speed at all quadrature points on all faces.
- z = mfem.Vector(A.Width())
- A.Mult(sol, z)
- max_char_speed = MPI.COMM_WORLD.allreduce(
- ex18_common.max_char_speed, op=MPI.MAX)
- ex18_common.max_char_speed = max_char_speed
- dt = cfl * hmin / ex18_common.max_char_speed / (2*order+1)
-
-# Integrate in time.
-done = False
-ti = 0
-while not done:
- dt_real = min(dt, t_final - t)
- t, dt_real = ode_solver.Step(sol, t, dt_real)
+ device.Print()
+
+ # 2. Read the mesh from the given mesh file. When the user does not provide
+ # mesh file, use the default mesh file for the problem.
+
+ mesh = EulerMesh(meshfile, problem)
+ dim = mesh.Dimension()
+ num_equation = dim + 2
+
+ # Refine the mesh to increase the resolution. In this example we do
+ # 'ser_ref_levels' of uniform refinement, where 'ser_ref_levels' is a
+ # command-line parameter.
+ for lev in range(ser_ref_levels):
+ mesh.UniformRefinement()
+
+ # Define a parallel mesh by a partitioning of the serial mesh. Refine this
+ # mesh further in parallel to increase the resolution. Once the parallel
+ # mesh is defined, the serial mesh can be deleted.
+ pmesh = mfem.ParMesh(MPI.COMM_WORLD, mesh)
+ del mesh
+
+ # Refine the mesh to increase the resolution. In this example we do
+ # 'par_ref_levels' of uniform refinement, where 'par_ref_levels' is a
+ # command-line parameter.
+ for lev in range(par_ref_levels):
+ pmesh.UniformRefinement()
+
+ # 3. Define the ODE solver used for time integration. Several explicit
+ # Runge-Kutta methods are available.
+ ode_solver = None
+ if ode_solver_type == 1:
+ ode_solver = mfem.ForwardEulerSolver()
+ elif ode_solver_type == 2:
+ ode_solver = mfem.RK2Solver(1.0)
+ elif ode_solver_type == 3:
+ ode_solver = mfem.RK3SSPSolver()
+ elif ode_solver_type == 4:
+ ode_solver = mfem.RK4Solver()
+ elif ode_solver_type == 6:
+ ode_solver = mfem.RK6Solver()
+ else:
+ print("Unknown ODE solver type: " + str(ode_solver_type))
+ exit
+
+ # 4. Define the discontinuous DG finite element space of the given
+ # polynomial order on the refined mesh.
+ fec = mfem.DG_FECollection(order, dim)
+ # Finite element space for a scalar (thermodynamic quantity)
+ fes = mfem.ParFiniteElementSpace(pmesh, fec)
+ # Finite element space for a mesh-dim vector quantity (momentum)
+ dfes = mfem.ParFiniteElementSpace(pmesh, fec, dim, mfem.Ordering.byNODES)
+ # Finite element space for all variables together (total thermodynamic state)
+ vfes = mfem.ParFiniteElementSpace(
+ pmesh, fec, num_equation, mfem.Ordering.byNODES)
+
+ assert fes.GetOrdering() == mfem.Ordering.byNODES, "Ordering must be byNODES"
+ glob_size = vfes.GlobalTrueVSize()
+ if myid == 0:
+ print("Number of unknowns: " + str(glob_size))
+
+ # 5. Define the initial conditions, save the corresponding mesh and grid
+ # functions to files. These can be opened with GLVis using:
+ # "glvis -np 4 -m euler-mesh -g euler-1-init" (for x-momentum).
+
+ # Initialize the state.
+ u0 = EulerInitialCondition(problem,
+ specific_heat_ratio,
+ gas_constant)
+ sol = mfem.ParGridFunction(vfes)
+ sol.ProjectCoefficient(u0)
+
+ # (Python note): GridFunction pointing to the subset of vector FES.
+ # sol is Vector with dim*fes.GetNDofs()
+ # Since sol.GetDataArray() returns numpy array pointing to the data, we make
+ # Vector from a sub-vector of the returned numpy array and pass it to GridFunction
+ # constructor.
+ mom = mfem.GridFunction(dfes, mfem.Vector(
+ sol.GetDataArray()[fes.GetNDofs():]))
+
+ # Output the initial solution.
+ smyid = '{:0>6d}'.format(myid)
+ pmesh.Print("euler-mesh."+smyid, 8)
+ for k in range(num_equation):
+ uk = mfem.ParGridFunction(fes, mfem.Vector(
+ sol.GetDataArray()[k*fes.GetNDofs():]))
+ sol_name = "euler-" + str(k) + "-init."+smyid
+ uk.Save(sol_name, 8)
+
+ # 6. Set up the nonlinear form with euler flux and numerical flux
+ flux = mfem.EulerFlux(dim, specific_heat_ratio)
+ numericalFlux = mfem.RusanovFlux(flux)
+ formIntegrator = mfem.HyperbolicFormIntegrator(
+ numericalFlux, IntOrderOffset)
+
+ euler = DGHyperbolicConservationLaws(vfes, formIntegrator,
+ preassembleWeakDivergence=preassembleWeakDiv)
+
+ # 7. Visualize momentum with its magnitude
+ if (visualization):
+ MPI.COMM_WORLD.Barrier()
+ sout = mfem.socketstream("localhost", 19916)
+ sout.precision(8)
+ sout << "parallel " << str(num_procs) << " " << str(myid) << "\n"
+ sout << "solution\n" << pmesh << mom
+ sout << "window_title 'momentum, t = 0'\n"
+ sout << "view 0 0\n" # view from top
+ sout << "keys jlm\n" # turn off perspective and light, show mesh
+ sout << "pause\n"
+ sout.flush()
+
+ if myid == 0:
+ print("GLVis visualization paused.")
+ print(" Press space (in the GLVis window) to resume it.")
+ # 8. Time integration
+ my_hmin = np.inf
if (cfl > 0):
+ my_hmin = min([pmesh.GetElementSize(i, 1)
+ for i in range(pmesh.GetNE())])
+
+ hmin = MPI.COMM_WORLD.allreduce(my_hmin, op=MPI.MIN)
+
+ # Find a safe dt, using a temporary vector. Calling Mult() computes the
+ # maximum char speed at all quadrature points on all faces (and all
+ # elements with -mf).
+ z = mfem.Vector(sol.Size())
+ euler.Mult(sol, z)
+
+ my_max_char_speed = euler.GetMaxCharSpeed()
max_char_speed = MPI.COMM_WORLD.allreduce(
- ex18_common.max_char_speed, op=MPI.MAX)
- ex18_common.max_char_speed = max_char_speed
- dt = cfl * hmin / ex18_common.max_char_speed / (2*order+1)
+ my_max_char_speed, op=MPI.MAX)
+
+ dt = cfl * hmin / max_char_speed / (2 * order + 1)
+
+ t = 0.0
+ euler.SetTime(t)
+ ode_solver.Init(euler)
+
+ # Integrate in time.
+ done = False
+ ti = 0
+ while not done:
+ dt_real = min(dt, t_final - t)
+ t, dt_real = ode_solver.Step(sol, t, dt_real)
+
+ if (cfl > 0):
+ my_max_char_speed = euler.GetMaxCharSpeed()
+ max_char_speed = MPI.COMM_WORLD.allreduce(
+ my_max_char_speed, op=MPI.MAX)
+
+ dt = cfl * hmin / max_char_speed / (2*order+1)
+
+ ti = ti+1
+ done = (t >= t_final - 1e-8*dt)
+ if (done or ti % vis_steps == 0):
+ if myid == 0:
+ print("time step: " + str(ti) + ", time: " + "{:g}".format(t))
+ if (visualization):
+ sout << "window_title 'momentum, t = " << "{:g}".format(
+ t) << "'\n"
+ sout << "parallel " << str(
+ num_procs) << " " << str(myid) << "\n"
+ sout << "solution\n" << pmesh << mom
+ sout.flush()
- ti = ti+1
- done = (t >= t_final - 1e-8*dt)
- if (done or ti % vis_steps == 0):
+ if myid == 0:
+ print("done")
+
+ # 9. Save the final solution. This output can be viewed later using GLVis:
+ # "glvis -np 4 -m euler-mesh-final -g euler-1-final" (for x-momentum).
+ pmesh.Print("euler-mesh-final."+smyid, 8)
+ for k in range(num_equation):
+ uk = mfem.ParGridFunction(fes, mfem.Vector(
+ sol.GetDataArray()[k*fes.GetNDofs():]))
+ sol_name = "euler-" + str(k) + "-final."+smyid
+ uk.Save(sol_name, 8)
+
+ # 10. Compute the L2 solution error summed for all components.
+ if True:
+ error = sol.ComputeLpError(2., u0)
if myid == 0:
- print("time step: " + str(ti) + ", time: " + "{:g}".format(t))
- if (visualization):
- sout.send_text("parallel " + str(num_procs) + " " + str(myid))
- sout.send_solution(pmesh, mom)
- sout.flush()
-
-if myid == 0:
- print("done")
-
-# 11. Save the final solution. This output can be viewed later using GLVis:
-# "glvis -np 4 -m vortex-mesh -g vortex-1-final".
-
-for k in range(num_equation):
- uk = mfem.ParGridFunction(fes, u_block.GetBlock(k).GetData())
- sol_name = "vortex-" + str(k) + "-final."+smyid
- uk.Save(sol_name, 8)
-
-# 12. Compute the L2 solution error summed for all components.
-# if (t_final == 2.0):
-if True:
- error = sol.ComputeLpError(2., u0)
+ print("Solution error: " + "{:g}".format(error))
+
+
+if __name__ == "__main__":
+
+ parser = ArgParser(description='Ex18p')
+ parser.add_argument('-m', '--mesh',
+ default='',
+ action='store', type=str,
+ help='Mesh file to use.')
+ parser.add_argument('-p', '--problem',
+ action='store', default=1, type=int,
+ help='Problem setup to use. See options in velocity_function().')
+ parser.add_argument('-rs', '--refine_serial',
+ action='store', default=0, type=int,
+ help="Number of times to refine the mesh uniformly before parallel.")
+ parser.add_argument('-rp', '--refine_parallel',
+ action='store', default=1, type=int,
+ help="Number of times to refine the mesh uniformly after parallel.")
+ parser.add_argument('-o', '--order',
+ action='store', default=3, type=int,
+ help="Finite element order (polynomial degree)")
+ parser.add_argument('-s', '--ode_solver',
+ action='store', default=4, type=int,
+ help="ODE solver: 1 - Forward Euler,\n\t" +
+ " 2 - RK2 SSP, 3 - RK3 SSP, 4 - RK4, 6 - RK6.")
+ parser.add_argument('-tf', '--t_final',
+ action='store', default=2.0, type=float,
+ help="Final time; start time is 0.")
+ parser.add_argument("-dt", "--time_step",
+ action='store', default=-0.01, type=float,
+ help="Time step.")
+ parser.add_argument('-c', '--cfl_number',
+ action='store', default=0.3, type=float,
+ help="CFL number for timestep calculation.")
+ parser.add_argument('-novis', '--no_visualization',
+ action='store_true', default=False,
+ help='Disable GLVis visualization')
+ parser.add_argument("-ea", "--element-assembly-divergence",
+ action='store_true', default=False,
+ help="Weak divergence assembly level\n" +
+ " ea - Element assembly with interpolated")
+ parser.add_argument("-mf", "--matrix-free-divergence",
+ action='store_true', default=False,
+ help="Weak divergence assembly level\n" +
+ " mf - Nonlinear assembly in matrix-free manner")
+ parser.add_argument('-vs', '--visualization-steps',
+ action='store', default=50, type=float,
+ help="Visualize every n-th timestep.")
+
+ args = parser.parse_args()
+
+ visualization = not args.no_visualization
+
+ if (not args.matrix_free_divergence and
+ not args.element_assembly_divergence):
+ args.element_assembly_divergence = True
+ args.matrix_free_divergence = False
+ preassembleWeakDiv = True
+
+ elif args.element_assembly_divergence:
+ args.matrix_free_divergence = False
+ preassembleWeakDiv = True
+
+ elif args.matrix_free_divergence:
+ args.element_assembly_divergence = False
+ preassembleWeakDiv = False
+
if myid == 0:
- print("Solution error: " + "{:g}".format(error))
+ parser.print_options(args)
+
+ run(problem=args.problem,
+ ser_ref_levels=args.refine_serial,
+ par_ref_levels=args.refine_parallel,
+ order=args.order,
+ ode_solver_type=args.ode_solver,
+ t_final=args.t_final,
+ dt=args.time_step,
+ cfl=args.cfl_number,
+ visualization=visualization,
+ vis_steps=args.visualization_steps,
+ preassembleWeakDiv=preassembleWeakDiv,
+ meshfile=args.mesh)
diff --git a/examples/ex19.py b/examples/ex19.py
index 91662728d..5fd869b4a 100644
--- a/examples/ex19.py
+++ b/examples/ex19.py
@@ -244,9 +244,9 @@ def SetOperator(self, op):
self.stiff_pcg.SetOperator(self.jacobian.GetBlock(0, 0))
-class GeneralResidualMonitor(mfem.IterativeSolverMonitor):
+class GeneralResidualMonitor(mfem.IterativeSolverController):
def __init__(self, prefix, print_level):
- mfem.IterativeSolverMonitor.__init__(self)
+ mfem.IterativeSolverController.__init__(self)
self.print_level = print_level
self.prefix = prefix
self.norm = 0
diff --git a/examples/ex19p.py b/examples/ex19p.py
index 4397c54fb..cd8080968 100644
--- a/examples/ex19p.py
+++ b/examples/ex19p.py
@@ -205,9 +205,9 @@ def ex19_main(args):
'''
-class GeneralResidualMonitor(mfem.IterativeSolverMonitor):
+class GeneralResidualMonitor(mfem.IterativeSolverController):
def __init__(self, prefix, print_level):
- mfem.IterativeSolverMonitor.__init__(self)
+ mfem.IterativeSolverController.__init__(self)
if myid == 0:
self.print_level = print_level
else:
diff --git a/examples/ex23.py b/examples/ex23.py
index e1d8fcb85..2ab4150fe 100644
--- a/examples/ex23.py
+++ b/examples/ex23.py
@@ -10,6 +10,9 @@
import numpy as np
from numpy import sin, cos, exp, sqrt, pi, abs, array, floor, log, sum
+mfem_version = (mfem.MFEM_VERSION_MAJOR*100 + mfem.MFEM_VERSION_MINOR*10+
+ mfem.MFEM_VERSION_PATCH)
+
def run(mesh_file="",
ref_levels=2,
@@ -30,7 +33,6 @@ def __init__(self, fespace, ess_bdr, speed):
self.ess_tdof_list = mfem.intArray()
- rel_tol = 1e-8
fespace.GetEssentialTrueDofs(ess_bdr, self.ess_tdof_list)
c2 = mfem.ConstantCoefficient(speed*speed)
@@ -38,20 +40,25 @@ def __init__(self, fespace, ess_bdr, speed):
K.AddDomainIntegrator(mfem.DiffusionIntegrator(c2))
K.Assemble()
- self.Kmat0 = mfem.SparseMatrix()
self.Kmat = mfem.SparseMatrix()
- dummy = mfem.intArray()
- K.FormSystemMatrix(dummy, self.Kmat0)
- K.FormSystemMatrix(self.ess_tdof_list, self.Kmat)
- self.K = K
-
self.Mmat = mfem.SparseMatrix()
+
M = mfem.BilinearForm(fespace)
M.AddDomainIntegrator(mfem.MassIntegrator())
M.Assemble()
+
+ if mfem_version < 471:
+ dummy = mfem.intArray()
+ self.Kmat0 = mfem.SparseMatrix()
+ K.FormSystemMatrix(dummy, self.Kmat0)
+ K.FormSystemMatrix(self.ess_tdof_list, self.Kmat)
M.FormSystemMatrix(self.ess_tdof_list, self.Mmat)
+
+ self.K = K
self.M = M
+ rel_tol = 1e-8
+
M_solver = mfem.CGSolver()
M_prec = mfem.DSmoother()
M_solver.iterative_mode = False
@@ -76,14 +83,19 @@ def __init__(self, fespace, ess_bdr, speed):
self.T_solver = T_solver
self.T = None
+ self.z = mfem.Vector(self.Height())
+
def Mult(self, u, du_dt, d2udt2):
# Compute:
# d2udt2 = M^{-1}*-K(u)
# for d2udt2
- z = mfem.Vector(u.Size())
- self.Kmat.Mult(u, z)
+ z = self.z
+ self.K.FullMult(u, z)
z.Neg() # z = -z
+
+ z.SetSubVector(self.ess_tdof_list, 0.0)
self.M_solver.Mult(z, d2udt2)
+ d2udt2.SetSubVector(self.ess_tdof_list, 0.0)
def ImplicitSolve(self, fac0, fac1, u, dudt, d2udt2):
# Solve the equation:
@@ -92,16 +104,21 @@ def ImplicitSolve(self, fac0, fac1, u, dudt, d2udt2):
if self.T is None:
self.T = mfem.Add(1.0, self.Mmat, fac0, self.Kmat)
self.T_solver.SetOperator(self.T)
- z = mfem.Vector(u.Size())
- self.Kmat0.Mult(u, z)
+
+ z = self.z
+ self.K.FullMult(u, z)
z.Neg()
+ z.SetSubVector(self.ess_tdof_list, 0.0)
# iterate over Array :D
- for j in self.ess_tdof_list:
- z[j] = 0.0
+ # for j in self.ess_tdof_list:
+ # z[j] = 0.0
self.T_solver.Mult(z, d2udt2)
+ if mfem_version >= 471:
+ d2udt2.SetSubVector(self.ess_tdof_list, 0.0)
+
def SetParameters(self, u):
self.T = None
@@ -169,7 +186,7 @@ def EvalValue(self, x):
if (dirichlet):
ess_bdr.Assign(1)
else:
- ess_bdr.Assigne(0)
+ ess_bdr.Assign(0)
oper = WaveOperator(fespace, ess_bdr, speed)
@@ -200,7 +217,7 @@ def EvalValue(self, x):
print("GLVis visualization disabled.")
else:
sout.precision(output.precision)
- sout << "solution\n" << mesh << dudt_gf
+ sout << "solution\n" << mesh << u_gf
sout << "pause\n"
sout.flush()
print(
diff --git a/examples/ex25.py b/examples/ex25.py
index eadbe5b85..f1ca092d0 100644
--- a/examples/ex25.py
+++ b/examples/ex25.py
@@ -13,16 +13,20 @@
'''
from numba import jit, types, carray
-import numba
-import numba_scipy
import os
import mfem.ser as mfem
from mfem.ser import intArray
from os.path import expanduser, join, dirname
import numpy as np
from numpy import sin, cos, exp, sqrt, pi
-import scipy.special
+try:
+ import numba
+except ImportError:
+ assert False, "Numba is required"
+
+from mfem.common.bessel import jv as jn
+from mfem.common.bessel import yv as yn
prob = ''
@@ -40,7 +44,6 @@ def run(meshfile="",
device = mfem.Device(device_config)
device.Print()
- print(prob)
# 3. Setup the mesh
if meshfile == '':
exact_known = True
@@ -529,8 +532,6 @@ def source(x, out):
def maxwell_solution(x, E, sdim):
- jn = scipy.special.jv
- yn = scipy.special.yn
# Initialize
for i in range(sdim):
E[i] = 0.0
@@ -686,7 +687,7 @@ def detJ_JT_J_inv_Im_f(x, D):
det *= dxs[i]
for i in range(dim):
D[i] = (det / (dxs[i]**2)).imag
- #print("imag", D)
+ # print("imag", D)
def detJ_JT_J_inv_abs_f(x, D):
diff --git a/examples/ex25p.py b/examples/ex25p.py
index a91972b1d..5b010a4e9 100644
--- a/examples/ex25p.py
+++ b/examples/ex25p.py
@@ -12,17 +12,22 @@
* calling JITed function from JITed coefficient
'''
+from mpi4py import MPI
+from mfem.common.bessel import yv as yn
+from mfem.common.bessel import jv as jn
from numba import jit, types, carray
-import numba
-import numba_scipy
import os
import mfem.par as mfem
from mfem.par import intArray
from os.path import expanduser, join, dirname
import numpy as np
from numpy import sin, cos, exp, sqrt, pi
-import scipy.special
-from mpi4py import MPI
+
+try:
+ import numba
+except ImportError:
+ assert False, "Numba is required"
+
num_procs = MPI.COMM_WORLD.size
myid = MPI.COMM_WORLD.rank
@@ -548,9 +553,6 @@ def source(x):
def maxwell_solution(x):
- jn = scipy.special.jv
- yn = scipy.special.yn
-
# Initialize
E = np.zeros(dim, dtype=np.complex128)
diff --git a/examples/ex27.py b/examples/ex27.py
index 7c2f5ddd9..d03252852 100644
--- a/examples/ex27.py
+++ b/examples/ex27.py
@@ -460,7 +460,7 @@ def run(order=1,
m.FormSystemMatrix(ess_tdof_list, M)
# 14. Compute the various boundary integrals.
- print("Verifying boundary conditions" +
+ print("Verifying boundary conditions\n" +
"=============================")
# Integrate the solution on the Dirichlet boundary and compare to the
diff --git a/examples/ex27p.py b/examples/ex27p.py
index d597e4867..41c9a2bbc 100644
--- a/examples/ex27p.py
+++ b/examples/ex27p.py
@@ -493,7 +493,7 @@ def run(order=1,
# 14. Compute the various boundary integrals.
if myid == 0:
- print("Verifying boundary conditions" +
+ print("Verifying boundary conditions\n" +
"=============================")
# Integrate the solution on the Dirichlet boundary and compare to the
diff --git a/examples/ex39.py b/examples/ex39.py
new file mode 100644
index 000000000..028ff47c4
--- /dev/null
+++ b/examples/ex39.py
@@ -0,0 +1,286 @@
+'''
+ MFEM example 39 (converted from ex39.cpp)
+
+ See c++ version in the MFEM library for more detail
+
+ Sample runs: python ex39.py
+ python ex39.py -ess "Southern Boundary"
+ python ex39.py -src Base
+
+ Description: This example code demonstrates the use of named attribute
+ sets in MFEM to specify material regions, boundary regions,
+ or source regions by name rather than attribute numbers. It
+ also demonstrates how new named attribute sets may be created
+ from arbitrary groupings of attribute numbers and used as a
+ convenient shorthand to refer to those groupings in other
+ portions of the application or through the command line.
+
+ The particular problem being solved here is nearly the same
+ as that in example 1 i.e. a simple finite element
+ discretization of the Laplace problem -Delta u = 1 with
+ homogeneous Dirichlet boundary conditions and, in this case,
+ an inhomogeneous diffusion coefficient. The diffusion
+ coefficient is given a small default value throughout the
+ domain which is increased by two separate amounts in two named
+ regions.
+
+ This example makes use of a specific input mesh, "compass.msh",
+ containing named domain and boundary regions generated by Gmsh
+ and stored in their "msh" format (version 2.2). This file
+ defines eight boundary regions corresponding to eight compass
+ headings; "ENE", "NNE", "NNW", "WSW", "SSW", "SSE", and "ESE".
+ It also defines nine domain regions; "Base", "N Even", "N Odd",
+ "W Even", "W Odd", "S Even", "S Odd", "E Even", and "E Odd".
+ These regions split the four compass pointers into two halves
+ each and also label the remaining elements as "Base". Starting
+ with these named regions we test the construction of named
+ sets as well as reading and writing these named groupings from
+ and to mesh files.
+
+ The example highlights the use of named attribute sets for
+ both subdomains and boundaries in different contexts as well
+ as basic methods to create named sets from existing attributes.
+
+'''
+import os
+from os.path import expanduser, join
+import numpy as np
+
+import mfem.ser as mfem
+
+
+def run(order=1,
+ meshfile='',
+ source_name='Rose Even',
+ ess_name='Boundary',
+ visualization=True):
+
+ # 2. Read the mesh from the given mesh file. We can handle triangular,
+ # quadrilateral, tetrahedral, hexahedral, surface and volume meshes with
+ # the same code.
+
+ mesh = mfem.Mesh(meshfile, 1, 1)
+ dim = mesh.Dimension()
+
+ # 3. Refine the mesh to increase the resolution. In this example we do
+ # 'ref_levels' of uniform refinement. We choose 'ref_levels' to be the
+ # largest number that gives a final mesh with no more than 50,000
+ # elements.
+ ref_levels = int(np.log(50000./mesh.GetNE())/np.log(2.)/dim)
+ for i in range(ref_levels):
+ mesh.UniformRefinement()
+
+ # 4a. Display attribute set names contained in the initial mesh
+ # GetAttributeSetNames returns Python set object.
+ attr_sets = mesh.attribute_sets
+ bdr_attr_sets = mesh.bdr_attribute_sets
+ names = attr_sets.GetAttributeSetNames()
+ print("Element Attribute Set Names: " +
+ ' '.join(['"' + x + '"' for x in names]))
+ names = bdr_attr_sets.GetAttributeSetNames()
+ print("Boundary Attribute Set Names: " +
+ ' '.join(['"' + x + '"' for x in names]))
+
+ # 4b. Define new regions based on existing attribute sets
+ Na = attr_sets.GetAttributeSet("N Even")
+ Nb = attr_sets.GetAttributeSet("N Odd")
+ Sa = attr_sets.GetAttributeSet("S Even")
+ Sb = attr_sets.GetAttributeSet("S Odd")
+ Ea = attr_sets.GetAttributeSet("E Even")
+ Eb = attr_sets.GetAttributeSet("E Odd")
+ Wa = attr_sets.GetAttributeSet("W Even")
+ Wb = attr_sets.GetAttributeSet("W Odd")
+
+ # Create a new set spanning the North point
+ attr_sets.SetAttributeSet("North", Na)
+ attr_sets.AddToAttributeSet("North", Nb)
+
+ # Create a new set spanning the South point
+ attr_sets.SetAttributeSet("South", Sa)
+ attr_sets.AddToAttributeSet("South", Sb)
+
+ # Create a new set spanning the East point
+ attr_sets.SetAttributeSet("East", Ea)
+ attr_sets.AddToAttributeSet("East", Eb)
+
+ # Create a new set spanning the West point
+ attr_sets.SetAttributeSet("West", Wa)
+ attr_sets.AddToAttributeSet("West", Wb)
+
+ # Create a new set consisting of the "a" sides of the compass rose
+ attr_sets.SetAttributeSet("Rose Even", Na)
+ attr_sets.AddToAttributeSet("Rose Even", Sa)
+ attr_sets.AddToAttributeSet("Rose Even", Ea)
+ attr_sets.AddToAttributeSet("Rose Even", Wa)
+
+ # Create a new set consisting of the "b" sides of the compass rose
+ attr_sets.SetAttributeSet("Rose Odd", Nb)
+ attr_sets.AddToAttributeSet("Rose Odd", Sb)
+ attr_sets.AddToAttributeSet("Rose Odd", Eb)
+ attr_sets.AddToAttributeSet("Rose Odd", Wb)
+
+ # Create a new set consisting of the full compass rose
+ Ra = attr_sets.GetAttributeSet("Rose Even")
+ Rb = attr_sets.GetAttributeSet("Rose Odd")
+ attr_sets.SetAttributeSet("Rose", Ra)
+ attr_sets.AddToAttributeSet("Rose", Rb)
+
+ # 4c. Define new boundary regions based on existing boundary attribute sets
+ NNE = bdr_attr_sets.GetAttributeSet("NNE")
+ NNW = bdr_attr_sets.GetAttributeSet("NNW")
+ ENE = bdr_attr_sets.GetAttributeSet("ENE")
+ ESE = bdr_attr_sets.GetAttributeSet("ESE")
+ SSE = bdr_attr_sets.GetAttributeSet("SSE")
+ SSW = bdr_attr_sets.GetAttributeSet("SSW")
+ WNW = bdr_attr_sets.GetAttributeSet("WNW")
+ WSW = bdr_attr_sets.GetAttributeSet("WSW")
+
+ bdr_attr_sets.SetAttributeSet("Northern Boundary", NNE)
+ bdr_attr_sets.AddToAttributeSet("Northern Boundary", NNW)
+
+ bdr_attr_sets.SetAttributeSet("Southern Boundary", SSE)
+ bdr_attr_sets.AddToAttributeSet("Southern Boundary", SSW)
+
+ bdr_attr_sets.SetAttributeSet("Eastern Boundary", ENE)
+ bdr_attr_sets.AddToAttributeSet("Eastern Boundary", ESE)
+
+ bdr_attr_sets.SetAttributeSet("Western Boundary", WNW)
+ bdr_attr_sets.AddToAttributeSet("Western Boundary", WSW)
+
+ bdr_attr_sets.SetAttributeSet("Boundary",
+ bdr_attr_sets.GetAttributeSet
+ ("Northern Boundary"))
+ bdr_attr_sets.AddToAttributeSet("Boundary",
+ bdr_attr_sets.GetAttributeSet
+ ("Southern Boundary"))
+ bdr_attr_sets.AddToAttributeSet("Boundary",
+ bdr_attr_sets.GetAttributeSet
+ ("Eastern Boundary"))
+ bdr_attr_sets.AddToAttributeSet("Boundary",
+ bdr_attr_sets.GetAttributeSet
+ ("Western Boundary"))
+
+ # 5. Define a finite element space on the mesh. Here we use continuous
+ # Lagrange finite elements of the specified order.
+
+ fec = mfem.H1_FECollection(order, dim)
+ fespace = mfem.FiniteElementSpace(mesh, fec)
+ print('Number of finite element unknowns: ' +
+ str(fespace.GetTrueVSize()))
+
+ # 6. Determine the list of true (i.e. conforming) essential boundary dofs.
+ # In this example, the boundary conditions are defined by marking all
+ # the boundary regions corresponding to the boundary attributes
+ # contained in the set named "ess_name" as essential (Dirichlet) and
+ # converting them to a list of true dofs.
+ ess_tdof_list = mfem.intArray()
+ if bdr_attr_sets.AttributeSetExists(ess_name):
+ ess_bdr_marker = bdr_attr_sets.GetAttributeSetMarker(ess_name)
+ fespace.GetEssentialTrueDofs(ess_bdr_marker, ess_tdof_list)
+
+ # 7. Set up the linear form b(.) which corresponds to the right-hand side of
+ # the FEM linear system, which in this case is (1_s,phi_i) where phi_i
+ # are the basis functions in fespace and 1_s is an indicator function
+ # equal to 1 on the region defined by the named set "source_name" and
+ # zero elsewhere.
+
+ source_marker = attr_sets.GetAttributeSetMarker(source_name)
+ b = mfem.LinearForm(fespace)
+ one = mfem.ConstantCoefficient(1.0)
+ b.AddDomainIntegrator(mfem.DomainLFIntegrator(one), source_marker)
+ b.Assemble()
+
+ # 8. Define the solution vector x as a finite element grid function
+ # corresponding to fespace. Initialize x with initial guess of zero,
+ # which satisfies the boundary conditions.
+ x = mfem.GridFunction(fespace)
+ x.Assign(0.0)
+
+ # 9. Set up the bilinear form a(.,.) on the finite element space
+ # corresponding to the Laplacian operator -Delta, by adding the
+ # Diffusion domain integrator.
+ a = mfem.BilinearForm(fespace)
+
+ defaultCoef = mfem.ConstantCoefficient(1.0e-6)
+ baseCoef = mfem.ConstantCoefficient(1.0)
+ roseCoef = mfem.ConstantCoefficient(2.0)
+
+ base_marker = attr_sets.GetAttributeSetMarker("Base")
+ rose_marker = attr_sets.GetAttributeSetMarker("Rose Even")
+
+ # Impose a very small diffusion coefficient across the entire mesh
+ a.AddDomainIntegrator(mfem.DiffusionIntegrator(defaultCoef))
+
+ # Impose an additional, stronger diffusion coefficient in select regions
+ a.AddDomainIntegrator(mfem.DiffusionIntegrator(baseCoef), base_marker)
+ a.AddDomainIntegrator(mfem.DiffusionIntegrator(roseCoef), rose_marker)
+
+ # 10. Assemble the bilinear form and the corresponding linear system,
+ # applying any necessary transformations.
+ a.Assemble()
+
+ A = mfem.SparseMatrix()
+ B = mfem.Vector()
+ X = mfem.Vector()
+ a.FormLinearSystem(ess_tdof_list, x, b, A, X, B)
+ print("Size of linear system: " + str(A.Height()))
+
+ # 11. Solve the system using PCG with symmetric Gauss-Seidel preconditioner.
+ M = mfem.GSSmoother(A)
+ mfem.PCG(A, M, B, X, 1, 800, 1e-12, 0.0)
+
+ # 12. Recover the solution as a finite element grid function.
+ a.RecoverFEMSolution(X, b, x)
+
+ # 13. Save the refined mesh and the solution. This output can be viewed
+ # later using GLVis: "glvis -m refined.mesh -g sol.gf".
+ mesh.Save('refined.mesh')
+ x.Save('sol.gf')
+
+
+ # 14. Send the solution by socket to a GLVis server.
+ if visualization:
+ sol_sock = mfem.socketstream("localhost", 19916)
+ sol_sock.precision(8)
+ sol_sock << "solution\n" << mesh << x << "keys Rjmm"
+ sol_sock.flush()
+
+
+if __name__ == "__main__":
+ from mfem.common.arg_parser import ArgParser
+
+ parser = ArgParser(description='Ex1 (Laplace Problem)')
+ parser.add_argument('-m', '--mesh',
+ default='compass.msh',
+ action='store', type=str,
+ help='Mesh file to use.')
+ parser.add_argument('-o', '--order',
+ action='store', default=1, type=int,
+ help='Finite element order (polynomial degree) or -1 for isoparametric space.')
+ parser.add_argument('-src', '--source-attr-name',
+ action='store', default='Rose Even', type=str,
+ help='Name of attribute set containing source.')
+ parser.add_argument('-ess', '--ess-attr-name',
+ action='store', default='Boundary', type=str,
+ help='Name of attribute set containing essential BC.')
+ parser.add_argument('-no-vis', '--no-visualization',
+ action='store_true',
+ default=False,
+ help='Disable or disable GLVis visualization')
+
+ args = parser.parse_args()
+ parser.print_options(args)
+
+ order = args.order
+ meshfile = expanduser(
+ join(os.path.dirname(__file__), '..', 'data', args.mesh))
+
+ visualization = not args.no_visualization
+ source_name = args.source_attr_name
+ ess_name = args.ess_attr_name
+
+ run(order=order,
+ meshfile=meshfile,
+ source_name=source_name,
+ ess_name=ess_name,
+ visualization=visualization)
diff --git a/examples/ex39p.py b/examples/ex39p.py
new file mode 100644
index 000000000..84115843e
--- /dev/null
+++ b/examples/ex39p.py
@@ -0,0 +1,311 @@
+'''
+ MFEM example 39 (converted from ex39.cpp)
+
+ See c++ version in the MFEM library for more detail
+
+ Sample runs: mpirun -np 4 python ex39p.py
+ mpirun -np 4 python ex39p.py -ess "Southern Boundary"
+ mpirun -np 4 python ex39p.py -src Base
+
+ Description: This example code demonstrates the use of named attribute
+ sets in MFEM to specify material regions, boundary regions,
+ or source regions by name rather than attribute numbers. It
+ also demonstrates how new named attribute sets may be created
+ from arbitrary groupings of attribute numbers and used as a
+ convenient shorthand to refer to those groupings in other
+ portions of the application or through the command line.
+
+ The particular problem being solved here is nearly the same
+ as that in example 1 i.e. a simple finite element
+ discretization of the Laplace problem -Delta u = 1 with
+ homogeneous Dirichlet boundary conditions and, in this case,
+ an inhomogeneous diffusion coefficient. The diffusion
+ coefficient is given a small default value throughout the
+ domain which is increased by two separate amounts in two named
+ regions.
+
+ This example makes use of a specific input mesh, "compass.msh",
+ containing named domain and boundary regions generated by Gmsh
+ and stored in their "msh" format (version 2.2). This file
+ defines eight boundary regions corresponding to eight compass
+ headings; "ENE", "NNE", "NNW", "WSW", "SSW", "SSE", and "ESE".
+ It also defines nine domain regions; "Base", "N Even", "N Odd",
+ "W Even", "W Odd", "S Even", "S Odd", "E Even", and "E Odd".
+ These regions split the four compass pointers into two halves
+ each and also label the remaining elements as "Base". Starting
+ with these named regions we test the construction of named
+ sets as well as reading and writing these named groupings from
+ and to mesh files.
+
+ The example highlights the use of named attribute sets for
+ both subdomains and boundaries in different contexts as well
+ as basic methods to create named sets from existing attributes.
+
+'''
+import os
+from os.path import expanduser, join
+import numpy as np
+
+import mfem.par as mfem
+
+from mpi4py import MPI
+num_procs = MPI.COMM_WORLD.size
+myid = MPI.COMM_WORLD.rank
+smyid = '.'+'{:0>6d}'.format(myid)
+
+def run(order=1,
+ meshfile='',
+ source_name='Rose Even',
+ ess_name='Boundary',
+ visualization=True):
+
+ # 3. Read the serial mesh from the given mesh file.
+ mesh = mfem.Mesh(meshfile, 1, 1)
+ dim = mesh.Dimension()
+
+ # 4. Refine the serial mesh on all processors to increase the resolution. In
+ # this example we do 'ref_levels' of uniform refinement. We choose
+ # 'ref_levels' to be the largest number that gives a final mesh with no
+ # more than 10,000 elements.
+
+ ref_levels = int(np.log(10000./mesh.GetNE())/np.log(2.)/dim)
+ for i in range(ref_levels):
+ mesh.UniformRefinement()
+
+ # 5. Define a parallel mesh by a partitioning of the serial mesh. Refine
+ # this mesh further in parallel to increase the resolution. Once the
+ # parallel mesh is defined, the serial mesh can be deleted.
+ pmesh = mfem.ParMesh(MPI.COMM_WORLD, mesh)
+ del mesh
+
+ par_ref_levels = 2
+ for l in range(par_ref_levels):
+ pmesh.UniformRefinement()
+
+ # 6a. Display attribute set names contained in the initial mesh
+ # GetAttributeSetNames returns Python set object.
+
+ attr_sets = pmesh.attribute_sets
+ bdr_attr_sets = pmesh.bdr_attribute_sets
+
+ if myid == 0:
+ names = attr_sets.GetAttributeSetNames()
+ print("Element Attribute Set Names: " +
+ ' '.join(['"' + x + '"' for x in names]))
+ names = bdr_attr_sets.GetAttributeSetNames()
+ print("Boundary Attribute Set Names: " +
+ ' '.join(['"' + x + '"' for x in names]))
+
+ # 6b. Define new regions based on existing attribute sets
+ Na = attr_sets.GetAttributeSet("N Even")
+ Nb = attr_sets.GetAttributeSet("N Odd")
+ Sa = attr_sets.GetAttributeSet("S Even")
+ Sb = attr_sets.GetAttributeSet("S Odd")
+ Ea = attr_sets.GetAttributeSet("E Even")
+ Eb = attr_sets.GetAttributeSet("E Odd")
+ Wa = attr_sets.GetAttributeSet("W Even")
+ Wb = attr_sets.GetAttributeSet("W Odd")
+
+ # Create a new set spanning the North point
+ attr_sets.SetAttributeSet("North", Na)
+ attr_sets.AddToAttributeSet("North", Nb)
+
+ # Create a new set spanning the South point
+ attr_sets.SetAttributeSet("South", Sa)
+ attr_sets.AddToAttributeSet("South", Sb)
+
+ # Create a new set spanning the East point
+ attr_sets.SetAttributeSet("East", Ea)
+ attr_sets.AddToAttributeSet("East", Eb)
+
+ # Create a new set spanning the West point
+ attr_sets.SetAttributeSet("West", Wa)
+ attr_sets.AddToAttributeSet("West", Wb)
+
+ # Create a new set consisting of the "a" sides of the compass rose
+ attr_sets.SetAttributeSet("Rose Even", Na)
+ attr_sets.AddToAttributeSet("Rose Even", Sa)
+ attr_sets.AddToAttributeSet("Rose Even", Ea)
+ attr_sets.AddToAttributeSet("Rose Even", Wa)
+
+ # Create a new set consisting of the "b" sides of the compass rose
+ attr_sets.SetAttributeSet("Rose Odd", Nb)
+ attr_sets.AddToAttributeSet("Rose Odd", Sb)
+ attr_sets.AddToAttributeSet("Rose Odd", Eb)
+ attr_sets.AddToAttributeSet("Rose Odd", Wb)
+
+ # Create a new set consisting of the full compass rose
+ Ra = attr_sets.GetAttributeSet("Rose Even")
+ Rb = attr_sets.GetAttributeSet("Rose Odd")
+ attr_sets.SetAttributeSet("Rose", Ra)
+ attr_sets.AddToAttributeSet("Rose", Rb)
+
+ # 6c. Define new boundary regions based on existing boundary attribute sets
+ NNE = bdr_attr_sets.GetAttributeSet("NNE")
+ NNW = bdr_attr_sets.GetAttributeSet("NNW")
+ ENE = bdr_attr_sets.GetAttributeSet("ENE")
+ ESE = bdr_attr_sets.GetAttributeSet("ESE")
+ SSE = bdr_attr_sets.GetAttributeSet("SSE")
+ SSW = bdr_attr_sets.GetAttributeSet("SSW")
+ WNW = bdr_attr_sets.GetAttributeSet("WNW")
+ WSW = bdr_attr_sets.GetAttributeSet("WSW")
+
+ bdr_attr_sets.SetAttributeSet("Northern Boundary", NNE)
+ bdr_attr_sets.AddToAttributeSet("Northern Boundary", NNW)
+
+ bdr_attr_sets.SetAttributeSet("Southern Boundary", SSE)
+ bdr_attr_sets.AddToAttributeSet("Southern Boundary", SSW)
+
+ bdr_attr_sets.SetAttributeSet("Eastern Boundary", ENE)
+ bdr_attr_sets.AddToAttributeSet("Eastern Boundary", ESE)
+
+ bdr_attr_sets.SetAttributeSet("Western Boundary", WNW)
+ bdr_attr_sets.AddToAttributeSet("Western Boundary", WSW)
+
+ bdr_attr_sets.SetAttributeSet("Boundary",
+ bdr_attr_sets.GetAttributeSet
+ ("Northern Boundary"))
+ bdr_attr_sets.AddToAttributeSet("Boundary",
+ bdr_attr_sets.GetAttributeSet
+ ("Southern Boundary"))
+ bdr_attr_sets.AddToAttributeSet("Boundary",
+ bdr_attr_sets.GetAttributeSet
+ ("Eastern Boundary"))
+ bdr_attr_sets.AddToAttributeSet("Boundary",
+ bdr_attr_sets.GetAttributeSet
+ ("Western Boundary"))
+
+ # 7. Define a parallel finite element space on the parallel mesh. Here we
+ # use continuous Lagrange finite elements of the specified order. If
+ # order < 1, we instead use an isoparametric/isogeometric space.
+ fec = mfem.H1_FECollection(order, dim)
+ fespace = mfem.ParFiniteElementSpace(pmesh, fec)
+ size = fespace.GlobalTrueVSize()
+ if myid == 0:
+ print('Number of finite element unknowns: ' + str(size))
+
+ # 8. Determine the list of true (i.e. parallel conforming) essential
+ # boundary dofs. In this example, the boundary conditions are defined
+ # by marking all the boundary regions corresponding to the boundary
+ # attributes contained in the set named "ess_name" as essential
+ # (Dirichlet) and converting them to a list of true dofs.
+ ess_tdof_list = mfem.intArray()
+ if bdr_attr_sets.AttributeSetExists(ess_name):
+ ess_bdr_marker = bdr_attr_sets.GetAttributeSetMarker(ess_name)
+ fespace.GetEssentialTrueDofs(ess_bdr_marker, ess_tdof_list)
+
+ # 9. Set up the parallel linear form b(.) which corresponds to the
+ # right-hand side of the FEM linear system, which in this case is
+ # (1_s,phi_i) where phi_i are the basis functions in fespace and 1_s
+ # is an indicator function equal to 1 on the region defined by the
+ # named set "source_name" and zero elsewhere.
+
+ source_marker = attr_sets.GetAttributeSetMarker(source_name)
+ b = mfem.ParLinearForm(fespace)
+ one = mfem.ConstantCoefficient(1.0)
+ b.AddDomainIntegrator(mfem.DomainLFIntegrator(one), source_marker)
+ b.Assemble()
+
+ # 10. Define the solution vector x as a parallel finite element grid
+ # function corresponding to fespace. Initialize x with initial guess of
+ # zero, which satisfies the boundary conditions.
+ x = mfem.ParGridFunction(fespace)
+ x.Assign(0.0)
+
+ # 11. Set up the parallel bilinear form a(.,.) on the finite element space
+ # corresponding to the Laplacian operator -Delta, by adding the
+ # Diffusion domain integrator.
+ a = mfem.ParBilinearForm(fespace)
+
+ defaultCoef = mfem.ConstantCoefficient(1.0e-6)
+ baseCoef = mfem.ConstantCoefficient(1.0)
+ roseCoef = mfem.ConstantCoefficient(2.0)
+
+ base_marker = attr_sets.GetAttributeSetMarker("Base")
+ rose_marker = attr_sets.GetAttributeSetMarker("Rose Even")
+
+ # Impose a very small diffusion coefficient across the entire mesh
+ a.AddDomainIntegrator(mfem.DiffusionIntegrator(defaultCoef))
+
+ # Impose an additional, stronger diffusion coefficient in select regions
+ a.AddDomainIntegrator(mfem.DiffusionIntegrator(baseCoef), base_marker)
+ a.AddDomainIntegrator(mfem.DiffusionIntegrator(roseCoef), rose_marker)
+
+ # 12. Assemble the bilinear form and the corresponding linear system,
+ # applying any necessary transformations.
+ a.Assemble()
+
+ A = mfem.HypreParMatrix()
+ B = mfem.Vector()
+ X = mfem.Vector()
+ a.FormLinearSystem(ess_tdof_list, x, b, A, X, B)
+
+ # 13. Solve the system using PCG with hypre's BoomerAMG preconditioner.
+ M = mfem.HypreBoomerAMG(A)
+ cg = mfem.CGSolver(MPI.COMM_WORLD)
+ cg.SetRelTol(1e-12)
+ cg.SetMaxIter(2000)
+ cg.SetPrintLevel(1)
+ cg.SetPreconditioner(M)
+ cg.SetOperator(A)
+ cg.Mult(B, X)
+
+ # 14. Recover the parallel grid function corresponding to X. This is the
+ # local finite element solution on each processor.
+ a.RecoverFEMSolution(X, b, x)
+
+ # 15. Save the refined mesh and the solution in parallel.
+ # This output can be viewed later using GLVis: "glvis -np -m mesh -g sol".
+ #
+ # Python note: In Python, suffix based on myid needs to be added explicitly.
+ pmesh.Print('mesh'+smyid)
+ x.Save('sol'+smyid)
+
+ # 16. Send the solution by socket to a GLVis server.
+ if visualization:
+ sol_sock = mfem.socketstream("localhost", 19916)
+ sol_sock << "parallel " << num_procs << " " << myid << "\n"
+ sol_sock.precision(8)
+ sol_sock << "solution\n" << pmesh << x << "keys Rjmm"
+ sol_sock.flush()
+
+
+if __name__ == "__main__":
+ from mfem.common.arg_parser import ArgParser
+
+ parser = ArgParser(description='Ex1 (Laplace Problem)')
+ parser.add_argument('-m', '--mesh',
+ default='compass.msh',
+ action='store', type=str,
+ help='Mesh file to use.')
+ parser.add_argument('-o', '--order',
+ action='store', default=1, type=int,
+ help='Finite element order (polynomial degree) or -1 for isoparametric space.')
+ parser.add_argument('-src', '--source-attr-name',
+ action='store', default='Rose Even', type=str,
+ help='Name of attribute set containing source.')
+ parser.add_argument('-ess', '--ess-attr-name',
+ action='store', default='Boundary', type=str,
+ help='Name of attribute set containing essential BC.')
+ parser.add_argument('-no-vis', '--no-visualization',
+ action='store_true',
+ default=False,
+ help='Disable or disable GLVis visualization')
+
+ args = parser.parse_args()
+ if myid == 0:
+ parser.print_options(args)
+
+ order = args.order
+ meshfile = expanduser(
+ join(os.path.dirname(__file__), '..', 'data', args.mesh))
+
+ visualization = not args.no_visualization
+ source_name = args.source_attr_name
+ ess_name = args.ess_attr_name
+
+ run(order=order,
+ meshfile=meshfile,
+ source_name=source_name,
+ ess_name=ess_name,
+ visualization=visualization)
diff --git a/examples/ex5p.py b/examples/ex5p.py
index 2ed895a28..51481f26d 100644
--- a/examples/ex5p.py
+++ b/examples/ex5p.py
@@ -306,7 +306,7 @@ def EvalValue(self, x):
u_sock << "parallel " << num_procs << " " << myid << "\n"
u_sock.precision(8)
u_sock << "solution\n" << pmesh << u << "window_title 'Velocity'\n"
- MPI.Barrier()
+ MPI.COMM_WORLD.Barrier()
p_sock = mfem.socketstream("localhost", 19916)
p_sock << "parallel " << num_procs << " " << myid << "\n"
p_sock.precision(8)
diff --git a/mfem/__init__.py b/mfem/__init__.py
index 0cb77b509..e59dcdbeb 100644
--- a/mfem/__init__.py
+++ b/mfem/__init__.py
@@ -20,5 +20,5 @@ def debug_print(message):
print(message)
-__version__ = '4.6.1.1'
+__version__ = '4.8.0rc1'
diff --git a/mfem/_par/array.i b/mfem/_par/array.i
index 95382e408..633aea487 100644
--- a/mfem/_par/array.i
+++ b/mfem/_par/array.i
@@ -86,6 +86,8 @@ XXXPTR_SIZE_IN(bool *data_, int asize, bool)
namespace mfem{
%pythonprepend Array::__setitem__ %{
i = int(i)
+ if hasattr(v, "thisown"):
+ v.thisown = False
%}
%pythonprepend Array::Append %{
if isinstance(args[0], list):
@@ -167,13 +169,62 @@ INSTANTIATE_ARRAY_BOOL
IGNORE_ARRAY_METHODS_PREMITIVE(unsigned int)
INSTANTIATE_ARRAY_NUMPYARRAY(uint, unsigned int, NPY_UINT) // 32bit
+/* Array< Array *> */
+IGNORE_ARRAY_METHODS(mfem::Array *)
+INSTANTIATE_ARRAY2(Array *, Array, intArray, 1)
+
/*
- for these Array2D, we instantiate it. But we dont extend it since, Array2D does not
- expose the interanl pointer to array1d.
+ Array2D:: Assign and data access
*/
+
+%extend mfem::Array2D{
+ void Assign(const T &a){
+ *self = a;
+ }
+ void Assign(const mfem::Array2D &a){
+ *self = a;
+ }
+
+ void __setitem__(PyObject* param, const T v) {
+ if (PyTuple_Check(param)) {
+ PyErr_Clear();
+ int i = PyInt_AsLong(PyTuple_GetItem(param, 0));
+ int j = PyInt_AsLong(PyTuple_GetItem(param, 1));
+
+ if (PyErr_Occurred()) {
+ PyErr_SetString(PyExc_ValueError, "Argument must be i, j");
+ return;
+ }
+ T *arr = self -> GetRow(i);
+ arr[j] = v;
+ }
+ }
+ T __getitem__(PyObject* param) {
+ if (PyTuple_Check(param)) {
+ PyErr_Clear();
+ int i = PyInt_AsLong(PyTuple_GetItem(param, 0));
+ int j = PyInt_AsLong(PyTuple_GetItem(param, 1));
+
+ if (PyErr_Occurred()) {
+ PyErr_SetString(PyExc_ValueError, "Argument must be i, j");
+ i = 0;
+ j = 0;
+ }
+ T *arr = self -> GetRow(i);
+ return arr[j];
+ }
+ }
+}
%template(intArray2D) mfem::Array2D;
%template(doubleArray2D) mfem::Array2D;
-/* Array< Array *> */
-IGNORE_ARRAY_METHODS(mfem::Array *)
-INSTANTIATE_ARRAY2(Array *, Array, intArray, 1)
+/* Array2D<* DenseMatrix>, Array2D<* SparseMatrix>, Array2D<* HypreParMatrix> */
+
+IGNORE_ARRAY_METHODS(mfem::DenseMatrix *)
+IGNORE_ARRAY_METHODS(mfem::SparseMatrix *)
+IGNORE_ARRAY_METHODS(mfem::HypreParMatrix *)
+
+%template(DenseMatrixArray2D) mfem::Array2D;
+%template(SparseMatrixArray2D) mfem::Array2D;
+%template(HypreParMatrixArray2D) mfem::Array2D;
+
diff --git a/mfem/_par/arrays_by_name.i b/mfem/_par/arrays_by_name.i
new file mode 100644
index 000000000..a2c7b9b9d
--- /dev/null
+++ b/mfem/_par/arrays_by_name.i
@@ -0,0 +1,31 @@
+%module(package="mfem._par") arrays_by_name
+%{
+#include "mfem.hpp"
+#include "numpy/arrayobject.h"
+#include "../common/io_stream.hpp"
+#include "../common/pyoperator.hpp"
+#include "../common/pycoefficient.hpp"
+#include "../common/pyintrules.hpp"
+%}
+
+%include "../common/existing_mfem_headers.i"
+
+#ifdef FILE_EXISTS_GENERAL_ARRAYS_BY_NAME
+%init %{
+import_array();
+%}
+
+%include "exception.i"
+%include "../common/exception.i"
+
+%import "array.i"
+%import "../common/io_stream_typemap.i"
+OSTREAM_TYPEMAP(std::ostream&)
+ISTREAM_TYPEMAP(std::istream&)
+
+%include "general/arrays_by_name.hpp"
+%ignore mfem::ArraysByName::begin;
+%ignore mfem::ArraysByName::end;
+
+%template(intArraysByName) mfem::ArraysByName;
+#endif
diff --git a/mfem/_par/attribute_sets.i b/mfem/_par/attribute_sets.i
new file mode 100644
index 000000000..1f130ccfd
--- /dev/null
+++ b/mfem/_par/attribute_sets.i
@@ -0,0 +1,41 @@
+%module(package="mfem._par") attribute_sets
+%{
+#include "mfem.hpp"
+#include "numpy/arrayobject.h"
+#include "../common/io_stream.hpp"
+#include "../common/pyoperator.hpp"
+#include "../common/pycoefficient.hpp"
+#include "../common/pyintrules.hpp"
+%}
+
+%include "../common/existing_mfem_headers.i"
+#ifdef FILE_EXISTS_MESH_ATTRIBUTE_SETS
+
+%init %{
+import_array();
+%}
+
+%include "exception.i"
+%include "../common/exception.i"
+
+%import "array.i"
+%import "arrays_by_name.i"
+%import "../common/io_stream_typemap.i"
+OSTREAM_TYPEMAP(std::ostream&)
+ISTREAM_TYPEMAP(std::istream&)
+
+//
+// AttributeSets::GetAttributeSetNames returns Python set
+//
+%typemap(out) std::set{
+ resultobj = PySet_New(NULL);
+ for (auto const &set_name : *(& $1)){
+ std::ostringstream name2;
+ name2 << set_name;
+ PySet_Add(resultobj, PyUnicode_FromString(name2.str().c_str()));
+ }
+}
+
+%include "mesh/attribute_sets.hpp"
+
+#endif
diff --git a/mfem/_par/bilinearform.i b/mfem/_par/bilinearform.i
index 078dce044..f237da2bf 100644
--- a/mfem/_par/bilinearform.i
+++ b/mfem/_par/bilinearform.i
@@ -2,6 +2,7 @@
%{
#include "fem/bilinearform.hpp"
#include "numpy/arrayobject.h"
+#include "../common/io_stream.hpp"
#include "../common/pyoperator.hpp"
#include "../common/pycoefficient.hpp"
#include "../common/pyintrules.hpp"
diff --git a/mfem/_par/bilininteg.i b/mfem/_par/bilininteg.i
index 2dbae46d7..2766e5a0c 100644
--- a/mfem/_par/bilininteg.i
+++ b/mfem/_par/bilininteg.i
@@ -40,6 +40,7 @@ import_array();
%ignore mfem::MassIntegrator::SetupPA;
+%include "../common/kernel_dispatch.i"
%include "fem/bilininteg.hpp"
%feature("director") mfem::PyBilinearFormIntegrator;
diff --git a/mfem/_par/coefficient.i b/mfem/_par/coefficient.i
index a93d8a084..9edcdc2f9 100644
--- a/mfem/_par/coefficient.i
+++ b/mfem/_par/coefficient.i
@@ -110,6 +110,23 @@ namespace mfem {
%include "../common/typemap_macros.i"
LIST_TO_MFEMOBJ_POINTERARRAY_IN(mfem::IntegrationRule const *irs[], mfem::IntegrationRule *, 0)
+LIST_TO_MFEMOBJ_ARRAY_IN(const mfem::Array & coefs, mfem::Coefficient *)
+LIST_TO_MFEMOBJ_ARRAY_IN(const mfem::Array & coefs, mfem::VectorCoefficient *)
+LIST_TO_MFEMOBJ_ARRAY_IN(const mfem::Array & coefs, mfem::MatrixCoefficient *)
+
+/* define CoefficientPtrArray, VectorCoefficientPtrArray, MatrixCoefficientPtrArray */
+%import "../common/array_listtuple_typemap.i"
+ARRAY_LISTTUPLE_INPUT_SWIGOBJ(mfem::Coefficient *, 1)
+ARRAY_LISTTUPLE_INPUT_SWIGOBJ(mfem::VectorCoefficient *, 1)
+ARRAY_LISTTUPLE_INPUT_SWIGOBJ(mfem::MatrixCoefficient *, 1)
+
+%import "../common/array_instantiation_macro.i"
+IGNORE_ARRAY_METHODS(mfem::Coefficient *)
+INSTANTIATE_ARRAY0(Coefficient *, Coefficient, 1)
+IGNORE_ARRAY_METHODS(mfem::VectorCoefficient *)
+INSTANTIATE_ARRAY0(VectorCoefficient *, VectorCoefficient, 1)
+IGNORE_ARRAY_METHODS(mfem::MatrixCoefficient *)
+INSTANTIATE_ARRAY0(MatrixCoefficient *, MatrixCoefficient, 1)
%include "fem/coefficient.hpp"
%include "../common/numba_coefficient.i"
diff --git a/mfem/_par/eltrans.i b/mfem/_par/eltrans.i
index 08b9f71a8..e41342d9a 100644
--- a/mfem/_par/eltrans.i
+++ b/mfem/_par/eltrans.i
@@ -22,6 +22,8 @@ import_array();
%import "intrules.i"
%import "geom.i"
+%import "../common/mfem_config.i"
+
%feature("shadow") mfem::ElementTransformation::Transform %{
def Transform(self, *args):
from .vector import Vector
@@ -35,5 +37,34 @@ def Transform(self, *args):
return _eltrans.ElementTransformation_Transform(self, *args)
%}
+%ignore mfem::ElementTransformation::TransformBack;
+%ignore mfem::IsoparametricTransformation::TransformBack;
+
+%include "../common/kernel_dispatch.i"
%include "fem/eltrans.hpp"
+//
+// special handling for TransformBack (this is because tol_0 is protected)
+//
+namespace mfem{
+ #ifdef MFEM_USE_DOUBLE
+ %extend IsoparametricTransformation{
+ virtual int _TransformBack(const Vector &pt, IntegrationPoint &ip,
+ const real_t phys_tol = 1e-15){
+ return self-> TransformBack(pt, ip, phys_tol);
+ }
+ }; //end of extend
+ #elif defined(MFEM_USE_SINGLE)
+ %extend IsoparametricTransformation{
+ virtual int _TransformBack(const Vector &pt, IntegrationPoint &ip,
+ const real_t phys_tol = 1e-7){
+ return self-> TransformBack(pt, ip, phys_tol);
+ }
+ }; //end of extend
+ #endif
+ } //end of namespace
+
+%pythoncode %{
+if hasattr(IsoparametricTransformation, "_TransformBack"):
+ IsoparametricTransformation.TransformBack = IsoparametricTransformation._TransformBack
+%}
diff --git a/mfem/_par/fe_base.i b/mfem/_par/fe_base.i
index dac6b233f..768fde43b 100644
--- a/mfem/_par/fe_base.i
+++ b/mfem/_par/fe_base.i
@@ -16,9 +16,16 @@ import_array();
%include "../common/typemap_macros.i"
%include "../common/exception.i"
- //%ignore FE;
+//forward declearation
+%inline %{
namespace mfem{
class FiniteElement;
}
+%}
+
+// this is to avoild extern breaks template
+namespace mfem{
+ %ignore poly1d;
+}
%include "fem/fe/fe_base.hpp"
diff --git a/mfem/_par/hyperbolic.i b/mfem/_par/hyperbolic.i
new file mode 100644
index 000000000..a56271576
--- /dev/null
+++ b/mfem/_par/hyperbolic.i
@@ -0,0 +1,36 @@
+%module(package="mfem._par") hyperbolic
+%feature("autodoc", "1");
+
+%{
+#include "mfem.hpp"
+#include "numpy/arrayobject.h"
+#include "../common/io_stream.hpp"
+#include "../common/pyoperator.hpp"
+#include "../common/pycoefficient.hpp"
+#include "../common/pyintrules.hpp"
+#include "../common/pynonlininteg.hpp"
+%}
+
+%include "../common/existing_mfem_headers.i"
+#ifdef FILE_EXISTS_FEM_HYPERBOLIC
+
+%init %{
+import_array();
+%}
+
+%include "exception.i"
+%include "std_string.i"
+%include "../common/exception.i"
+
+%import "array.i"
+%import "vector.i"
+%import "densemat.i"
+%import "eltrans.i"
+%import "nonlininteg.i"
+
+%include "fem/hyperbolic.hpp"
+
+#endif
+
+
+
diff --git a/mfem/_par/hypre.i b/mfem/_par/hypre.i
index 617d719b4..805932ddf 100644
--- a/mfem/_par/hypre.i
+++ b/mfem/_par/hypre.i
@@ -66,7 +66,7 @@ bool is_HYPRE_USING_CUDA(){
HYPRE_Int *col);
*/
-%typemap(in) (double *data_, HYPRE_BigInt *col)(PyArrayObject * tmp_arr1_ = NULL, PyArrayObject * tmp_arr2_ = NULL){
+%typemap(in) (mfem::real_t *data_, HYPRE_BigInt *col)(PyArrayObject * tmp_arr1_ = NULL, PyArrayObject * tmp_arr2_ = NULL){
//HypreParVec constructer requires outside object alive
// We keep reference to such outside numpy array in ProxyClass
tmp_arr1_ = (PyArrayObject *)PyList_GetItem($input,0);
@@ -75,9 +75,9 @@ bool is_HYPRE_USING_CUDA(){
$1 = (double *) PyArray_DATA(tmp_arr1_);
$2 = (HYPRE_BigInt *) PyArray_DATA(tmp_arr2_);
}
-%typemap(freearg) (double *data_, HYPRE_BigInt *col){
+%typemap(freearg) (mfem::real_t *data_, HYPRE_BigInt *col){
}
-%typemap(typecheck )(double *data_, HYPRE_BigInt *col){
+%typemap(typecheck )(mfem::real_t *data_, HYPRE_BigInt *col){
/* check if list of 2 numpy array or not */
if (!PyList_Check($input)) $1 = 0;
else {
@@ -103,7 +103,7 @@ bool is_HYPRE_USING_CUDA(){
%typemap(in) (int *I,
HYPRE_BigInt *J,
- double *data,
+ mfem::real_t *data,
HYPRE_BigInt *rows,
HYPRE_BigInt *cols)
(PyArrayObject *tmp_arr1_ = NULL,
@@ -131,7 +131,7 @@ bool is_HYPRE_USING_CUDA(){
}
}
%typemap(freearg) (int *I, HYPRE_BigInt *J,
- double *data, HYPRE_BigInt *rows, HYPRE_BigInt *cols){
+ mfem::real_t *data, HYPRE_BigInt *rows, HYPRE_BigInt *cols){
Py_XDECREF(tmp_arr1_$argnum);
Py_XDECREF(tmp_arr2_$argnum);
Py_XDECREF(tmp_arr3_$argnum);
@@ -142,7 +142,7 @@ bool is_HYPRE_USING_CUDA(){
}
%typemap(typecheck ) (int *I, HYPRE_BigInt *J,
- double *data, HYPRE_BigInt *rows,
+ mfem::real_t *data, HYPRE_BigInt *rows,
HYPRE_BigInt *cols){
/* check if list of 5 numpy array or not */
if (!PyList_Check($input)) $1 = 0;
diff --git a/mfem/_par/mesh.i b/mfem/_par/mesh.i
index 9abaead72..9f2cf0dc6 100644
--- a/mfem/_par/mesh.i
+++ b/mfem/_par/mesh.i
@@ -39,6 +39,7 @@ import_array();
%import "matrix.i"
%import "array.i"
+%import "attribute_sets.i"
%import "sort_pairs.i"
%import "ncmesh.i"
%import "vector.i"
@@ -217,30 +218,34 @@ def GetFaceElements(self, Face):
return Elem1.value(), Elem2.value()
%}
%feature("shadow") mfem::Mesh::GetElementTransformation %{
-def GetElementTransformation(self, i):
+def GetElementTransformation(self, i, Tr=None):
from mfem.par import IsoparametricTransformation
- Tr = IsoparametricTransformation()
+ if Tr is None:
+ Tr = IsoparametricTransformation()
$action(self, i, Tr)
return Tr
%}
%feature("shadow") mfem::Mesh::GetBdrElementTransformation %{
-def GetBdrElementTransformation(self, i):
+def GetBdrElementTransformation(self, i, Tr=None):
from mfem.par import IsoparametricTransformation
- Tr = IsoparametricTransformation()
+ if Tr is None:
+ Tr = IsoparametricTransformation()
$action(self, i, Tr)
return Tr
%}
%feature("shadow") mfem::Mesh::GetFaceTransformation %{
-def GetFaceTransformation(self, i):
+def GetFaceTransformation(self, i, Tr=None):
from mfem.par import IsoparametricTransformation
- Tr = IsoparametricTransformation()
+ if Tr is None:
+ Tr = IsoparametricTransformation()
$action(self, i, Tr)
return Tr
%}
%feature("shadow") mfem::Mesh::GetEdgeTransformation %{
-def GetEdgeTransformation(self, i):
+def GetEdgeTransformation(self, i, Tr=None):
from mfem.par import IsoparametricTransformation
- Tr = IsoparametricTransformation()
+ if Tr is None:
+ Tr = IsoparametricTransformation()
$action(self, i, Tr)
return Tr
%}
@@ -304,6 +309,10 @@ def CartesianPartitioning(self, nxyz, return_list=False):
%newobject mfem::Mesh::GetFaceToElementTable;
%newobject mfem::Mesh::GetVertexToElementTable;
+%immutable mfem::MeshPart::mesh;
+%immutable mfem::MeshPart::nodal_fes;
+%immutable mfem::MeshPart::nodes;
+
%include "mesh/mesh.hpp"
%mutable;
diff --git a/mfem/_par/ncmesh.i b/mfem/_par/ncmesh.i
index 98404bed6..d43135fe1 100644
--- a/mfem/_par/ncmesh.i
+++ b/mfem/_par/ncmesh.i
@@ -35,6 +35,12 @@ ARRAY_LISTTUPLE_INPUT_SWIGOBJ(mfem::Refinement, 0)
XXXPTR_SIZE_IN(mfem::Refinement *data_, int asize, mfem::Refinement)
%immutable embeddings;
+
+%ignore mfem::NCMesh::FindFaceNodes(const Face &fa) const;
+%ignore mfem::NCMesh::GetElement(int i) const;
+%ignore mfem::NCMesh::GetFace(int i) const;
+%ignore mfem::NCMesh::GetNode(int i) const;
+
%include "mesh/ncmesh.hpp"
/*
diff --git a/mfem/_par/pbilinearform.i b/mfem/_par/pbilinearform.i
index 8a04358b4..191f0b9e3 100644
--- a/mfem/_par/pbilinearform.i
+++ b/mfem/_par/pbilinearform.i
@@ -23,6 +23,7 @@ import_array();
%import "handle.i"
%import "bilinearform.i"
%import "pfespace.i"
+%import "pgridfunc.i"
%import "hypre.i"
%import "../common/exception.i"
diff --git a/mfem/_par/quadinterpolator.i b/mfem/_par/quadinterpolator.i
index 383d03f36..650bd3956 100644
--- a/mfem/_par/quadinterpolator.i
+++ b/mfem/_par/quadinterpolator.i
@@ -25,6 +25,7 @@ import_array();
%import "../common/numpy_int_typemap.i"
+%include "../common/kernel_dispatch.i"
%include "fem/quadinterpolator.hpp"
#endif
diff --git a/mfem/_par/setup.py b/mfem/_par/setup.py
index 4e241d5c7..f879469ed 100644
--- a/mfem/_par/setup.py
+++ b/mfem/_par/setup.py
@@ -3,12 +3,17 @@
"""
setup.py file for SWIG example
"""
-from distutils.core import Extension, setup
-import numpy
-import os
+
import sys
+import os
+import numpy
+
print('building paralel version')
+# this remove *.py in this directory to be imported from setuptools
+sys.path.remove(os.path.abspath(os.path.dirname(sys.argv[0])))
+from distutils.core import Extension, setup
+
# first load variables from PyMFEM_ROOT/setup_local.py
@@ -121,7 +126,9 @@ def get_extensions():
"tmop", "tmop_amr", "tmop_tools", "qspace", "qfunction",
"quadinterpolator", "quadinterpolator_face",
"submesh", "transfermap", "staticcond","sidredatacollection",
- "psubmesh", "ptransfermap", "enzyme"]
+ "psubmesh", "ptransfermap", "enzyme",
+ "attribute_sets", "arrays_by_name",
+ "hyperbolic"]
if add_pumi == '1':
from setup_local import puminc, pumilib
diff --git a/mfem/_par/solvers.i b/mfem/_par/solvers.i
index 49e46c4e5..027fc6014 100644
--- a/mfem/_par/solvers.i
+++ b/mfem/_par/solvers.i
@@ -31,10 +31,16 @@ import_array();
%import "hypre.i"
%import "../common/exception.i"
-%ignore mfem::IterativeSolverMonitor::SetIterativeSolver;
%feature("director") mfem::IterativeSolverMonitor;
+%feature("director") mfem::IterativeSolverController;
%feature("director") mfem::PyIterativeSolver;
+// Forward declaration
+%inline %{
+ namespace mfem{
+ class IterativeSolver;
+ }
+%}
%include "linalg/solvers.hpp"
%include "../common/pysolvers.hpp"
diff --git a/mfem/_par/sparsemat.i b/mfem/_par/sparsemat.i
index ebbd9f56e..1a5c97f77 100644
--- a/mfem/_par/sparsemat.i
+++ b/mfem/_par/sparsemat.i
@@ -20,6 +20,8 @@ using namespace mfem;
import_array();
%}
+%import "../common/mfem_config.i"
+
%include "exception.i"
%import "array.i"
%import "mem_manager.i"
@@ -100,7 +102,7 @@ if len(args) == 1 and isinstance(args[0], csr_matrix):
SparseMatrix(int *i, int *j, double *data, int m, int n);
allows to use numpy array to call this
*/
-%typemap(in) (int *i, int *j, double *data, int m, int n)
+%typemap(in) (int *i, int *j, mfem::real_t *data, int m, int n)
(PyArrayObject *tmp_arr1_ = NULL,
PyArrayObject *tmp_arr2_ = NULL,
PyArrayObject *tmp_arr3_ = NULL,
@@ -121,13 +123,13 @@ if len(args) == 1 and isinstance(args[0], csr_matrix):
//
PyArray_CLEARFLAGS(tmp_arr3_, NPY_ARRAY_OWNDATA);
}
-%typemap(freearg) (int *i, int *j, double *data, int m, int n){
+%typemap(freearg) (int *i, int *j, mfem::real_t *data, int m, int n){
//Py_XDECREF(tmp_arr1_$argnum); Dont do this.. We set OwnsGraph and OwnsData to Fase in Python
//Py_XDECREF(tmp_arr2_$argnum);
//Py_XDECREF(tmp_arr3_$argnum);
}
-%typemap(typecheck ) (int *i, int *j, double *data, int m, int n){
+%typemap(typecheck ) (int *i, int *j, mfem::real_t *data, int m, int n){
/* check if list of 5 numpy array or not */
if (!PyList_Check($input)) $1 = 0;
else {
diff --git a/mfem/_par/transfer.i b/mfem/_par/transfer.i
index 969950d12..952422569 100644
--- a/mfem/_par/transfer.i
+++ b/mfem/_par/transfer.i
@@ -17,6 +17,7 @@ import_array();
%include "exception.i"
%import "operators.i"
+%import "device.i"
%import "fespace.i"
%import "pfespace.i"
%include "../common/exception.i"
diff --git a/mfem/_ser/array.i b/mfem/_ser/array.i
index 5d9f8e2aa..64bf6f4c4 100644
--- a/mfem/_ser/array.i
+++ b/mfem/_ser/array.i
@@ -40,17 +40,6 @@ XXXPTR_SIZE_IN(int *data_, int asize, int)
XXXPTR_SIZE_IN(double *data_, int asize, double)
XXXPTR_SIZE_IN(bool *data_, int asize, bool)
-%pythonappend mfem::Array::Array %{
- if len(args) == 1 and isinstance(args[0], list):
- if (len(args[0]) == 2 and hasattr(args[0][0], 'disown') and
- not hasattr(args[0][1], 'disown')):
- ## first element is SwigObject, like
- ## We do not own data in this case.
- pass
- else:
- self.MakeDataOwner()
-%}
-
%ignore mfem::Array::operator[];
%ignore mfem::Array2D::operator[];
%ignore mfem::BlockArray::operator[];
@@ -92,8 +81,20 @@ XXXPTR_SIZE_IN(bool *data_, int asize, bool)
};
namespace mfem{
+%pythonappend Array::Array %{
+ if len(args) == 1 and isinstance(args[0], list):
+ if (len(args[0]) == 2 and hasattr(args[0][0], 'disown') and
+ not hasattr(args[0][1], 'disown')):
+ ## first element is SwigObject, like
+ ## We do not own data in this case.
+ pass
+ else:
+ self.MakeDataOwner()
+%}
%pythonprepend Array::__setitem__ %{
i = int(i)
+ if hasattr(v, "thisown"):
+ v.thisown = False
%}
%pythonprepend Array::Append %{
if isinstance(args[0], list):
@@ -179,14 +180,60 @@ INSTANTIATE_ARRAY_BOOL
IGNORE_ARRAY_METHODS_PREMITIVE(unsigned int)
INSTANTIATE_ARRAY_NUMPYARRAY(uint, unsigned int, NPY_UINT) // 32bit
+/* Array< Array *> */
+IGNORE_ARRAY_METHODS(mfem::Array *)
+INSTANTIATE_ARRAY2(Array *, Array, intArray, 1)
+
/*
- for these Array2D, we instantiate it. But we dont extend it since, Array2D does not
- expose the interanl pointer to array1d.
+ Array2D:: Assign and data access
*/
+
+%extend mfem::Array2D{
+ void Assign(const T &a){
+ *self = a;
+ }
+ void Assign(const mfem::Array2D &a){
+ *self = a;
+ }
+
+ void __setitem__(PyObject* param, const T v) {
+ if (PyTuple_Check(param)) {
+ PyErr_Clear();
+ int i = PyInt_AsLong(PyTuple_GetItem(param, 0));
+ int j = PyInt_AsLong(PyTuple_GetItem(param, 1));
+
+ if (PyErr_Occurred()) {
+ PyErr_SetString(PyExc_ValueError, "Argument must be i, j");
+ return;
+ }
+ T *arr = self -> GetRow(i);
+ arr[j] = v;
+ }
+ }
+ T __getitem__(PyObject* param) {
+ if (PyTuple_Check(param)) {
+ PyErr_Clear();
+ int i = PyInt_AsLong(PyTuple_GetItem(param, 0));
+ int j = PyInt_AsLong(PyTuple_GetItem(param, 1));
+
+ if (PyErr_Occurred()) {
+ PyErr_SetString(PyExc_ValueError, "Argument must be i, j");
+ i = 0;
+ j = 0;
+ }
+ T *arr = self -> GetRow(i);
+ return arr[j];
+ }
+ }
+}
+
%template(intArray2D) mfem::Array2D;
%template(doubleArray2D) mfem::Array2D;
-/* Array< Array *> */
-IGNORE_ARRAY_METHODS(mfem::Array *)
-INSTANTIATE_ARRAY2(Array *, Array, intArray, 1)
+/* Array2D<* DenseMatrix>, Array2D<* SparseMatrix>, Array2D<* HypreParMatrix> */
+
+IGNORE_ARRAY_METHODS(mfem::DenseMatrix *)
+IGNORE_ARRAY_METHODS(mfem::SparseMatrix *)
+%template(DenseMatrixArray2D) mfem::Array2D;
+%template(SparseMatrixArray2D) mfem::Array2D;
diff --git a/mfem/_ser/arrays_by_name.i b/mfem/_ser/arrays_by_name.i
new file mode 100644
index 000000000..0ba9c95d3
--- /dev/null
+++ b/mfem/_ser/arrays_by_name.i
@@ -0,0 +1,31 @@
+%module(package="mfem._ser") arrays_by_name
+%{
+#include "mfem.hpp"
+#include "numpy/arrayobject.h"
+#include "../common/io_stream.hpp"
+#include "../common/pyoperator.hpp"
+#include "../common/pycoefficient.hpp"
+#include "../common/pyintrules.hpp"
+%}
+
+%include "../common/existing_mfem_headers.i"
+
+#ifdef FILE_EXISTS_GENERAL_ARRAYS_BY_NAME
+%init %{
+import_array();
+%}
+
+%include "exception.i"
+%include "../common/exception.i"
+
+%import "array.i"
+%import "../common/io_stream_typemap.i"
+OSTREAM_TYPEMAP(std::ostream&)
+ISTREAM_TYPEMAP(std::istream&)
+
+%include "general/arrays_by_name.hpp"
+%ignore mfem::ArraysByName::begin;
+%ignore mfem::ArraysByName::end;
+
+%template(intArraysByName) mfem::ArraysByName;
+#endif
diff --git a/mfem/_ser/attribute_sets.i b/mfem/_ser/attribute_sets.i
new file mode 100644
index 000000000..b9fd738fe
--- /dev/null
+++ b/mfem/_ser/attribute_sets.i
@@ -0,0 +1,47 @@
+%module(package="mfem._ser") attribute_sets
+%feature("autodoc", "1");
+
+%{
+#include "mfem.hpp"
+#include "numpy/arrayobject.h"
+#include "../common/io_stream.hpp"
+#include "../common/pyoperator.hpp"
+#include "../common/pycoefficient.hpp"
+#include "../common/pyintrules.hpp"
+%}
+
+%include "../common/existing_mfem_headers.i"
+#ifdef FILE_EXISTS_MESH_ATTRIBUTE_SETS
+
+%init %{
+import_array();
+%}
+
+%include "exception.i"
+%include "std_string.i"
+%include "../common/exception.i"
+
+%import "array.i"
+%import "arrays_by_name.i"
+%import "../common/io_stream_typemap.i"
+OSTREAM_TYPEMAP(std::ostream&)
+ISTREAM_TYPEMAP(std::istream&)
+
+//
+// AttributeSets::GetAttributeSetNames returns Python set
+//
+%typemap(out) std::set{
+ resultobj = PySet_New(NULL);
+ for (auto const &set_name : *(& $1)){
+ std::ostringstream name2;
+ name2 << set_name;
+ PySet_Add(resultobj, PyUnicode_FromString(name2.str().c_str()));
+ }
+}
+
+%include "mesh/attribute_sets.hpp"
+
+#endif
+
+
+
diff --git a/mfem/_ser/bilinearform.i b/mfem/_ser/bilinearform.i
index c2876bc9c..d30da958c 100644
--- a/mfem/_ser/bilinearform.i
+++ b/mfem/_ser/bilinearform.i
@@ -2,6 +2,7 @@
%{
#include "fem/bilinearform.hpp"
#include "numpy/arrayobject.h"
+#include "../common/io_stream.hpp"
#include "../common/pyoperator.hpp"
#include "../common/pycoefficient.hpp"
#include "../common/pyintrules.hpp"
diff --git a/mfem/_ser/bilininteg.i b/mfem/_ser/bilininteg.i
index 1acb4445c..8d549a2ac 100644
--- a/mfem/_ser/bilininteg.i
+++ b/mfem/_ser/bilininteg.i
@@ -38,6 +38,7 @@ import_array();
%ignore mfem::MassIntegrator::SetupPA;
+%include "../common/kernel_dispatch.i"
%include "fem/bilininteg.hpp"
%feature("director") mfem::PyBilinearFormIntegrator;
diff --git a/mfem/_ser/coefficient.i b/mfem/_ser/coefficient.i
index 7c37066df..6809e2660 100644
--- a/mfem/_ser/coefficient.i
+++ b/mfem/_ser/coefficient.i
@@ -110,6 +110,23 @@ namespace mfem {
*/
%include "../common/typemap_macros.i"
LIST_TO_MFEMOBJ_POINTERARRAY_IN(mfem::IntegrationRule const *irs[], mfem::IntegrationRule *, 0)
+LIST_TO_MFEMOBJ_ARRAY_IN(const mfem::Array & coefs, mfem::Coefficient *)
+LIST_TO_MFEMOBJ_ARRAY_IN(const mfem::Array & coefs, mfem::VectorCoefficient *)
+LIST_TO_MFEMOBJ_ARRAY_IN(const mfem::Array & coefs, mfem::MatrixCoefficient *)
+
+/* define CoefficientPtrArray, VectorCoefficientPtrArray, MatrixCoefficientPtrArray */
+%import "../common/array_listtuple_typemap.i"
+ARRAY_LISTTUPLE_INPUT_SWIGOBJ(mfem::Coefficient *, 1)
+ARRAY_LISTTUPLE_INPUT_SWIGOBJ(mfem::VectorCoefficient *, 1)
+ARRAY_LISTTUPLE_INPUT_SWIGOBJ(mfem::MatrixCoefficient *, 1)
+
+%import "../common/array_instantiation_macro.i"
+IGNORE_ARRAY_METHODS(mfem::Coefficient *)
+INSTANTIATE_ARRAY0(Coefficient *, Coefficient, 1)
+IGNORE_ARRAY_METHODS(mfem::VectorCoefficient *)
+INSTANTIATE_ARRAY0(VectorCoefficient *, VectorCoefficient, 1)
+IGNORE_ARRAY_METHODS(mfem::MatrixCoefficient *)
+INSTANTIATE_ARRAY0(MatrixCoefficient *, MatrixCoefficient, 1)
%include "fem/coefficient.hpp"
%include "../common/numba_coefficient.i"
diff --git a/mfem/_ser/eltrans.i b/mfem/_ser/eltrans.i
index caeaf5747..9e6aea8b9 100644
--- a/mfem/_ser/eltrans.i
+++ b/mfem/_ser/eltrans.i
@@ -22,6 +22,7 @@ import_array();
%import "intrules.i"
%import "geom.i"
%import "../common/exception.i"
+%import "../common/mfem_config.i"
%feature("shadow") mfem::ElementTransformation::Transform %{
def Transform(self, *args):
@@ -39,5 +40,37 @@ def Transform(self, *args):
%include "../common/deprecation.i"
DEPRECATED_METHOD(mfem::IsoparametricTransformation::FinalizeTransformation())
+
+%ignore mfem::ElementTransformation::TransformBack;
+%ignore mfem::IsoparametricTransformation::TransformBack;
+
+%include "../common/kernel_dispatch.i"
%include "fem/eltrans.hpp"
+//
+// special handling for TransformBack (this is because tol_0 is protected)
+//
+namespace mfem{
+ #ifdef MFEM_USE_DOUBLE
+ %extend IsoparametricTransformation{
+ virtual int _TransformBack(const Vector &pt, IntegrationPoint &ip,
+ const real_t phys_tol = 1e-15){
+ return self-> TransformBack(pt, ip, phys_tol);
+ }
+ }; //end of extend
+ #elif defined(MFEM_USE_SINGLE)
+ %extend IsoparametricTransformation{
+ virtual int _TransformBack(const Vector &pt, IntegrationPoint &ip,
+ const real_t phys_tol = 1e-7){
+ return self-> TransformBack(pt, ip, phys_tol);
+ }
+ }; //end of extend
+ #endif
+ } //end of namespace
+
+%pythoncode %{
+if hasattr(IsoparametricTransformation, "_TransformBack"):
+ IsoparametricTransformation.TransformBack = IsoparametricTransformation._TransformBack
+%}
+
+
diff --git a/mfem/_ser/fe_base.i b/mfem/_ser/fe_base.i
index aa26bc873..1d3800456 100644
--- a/mfem/_ser/fe_base.i
+++ b/mfem/_ser/fe_base.i
@@ -16,9 +16,16 @@ import_array();
%include "../common/typemap_macros.i"
%include "../common/exception.i"
- //%ignore FE;
+//forward declearation
+%inline %{
namespace mfem{
class FiniteElement;
}
+%}
+// this is to avoild extern breaks template
+namespace mfem{
+ %ignore poly1d;
+}
%include "fem/fe/fe_base.hpp"
+
diff --git a/mfem/_ser/hyperbolic.i b/mfem/_ser/hyperbolic.i
new file mode 100644
index 000000000..9047fab0a
--- /dev/null
+++ b/mfem/_ser/hyperbolic.i
@@ -0,0 +1,36 @@
+%module(package="mfem._ser") hyperbolic
+%feature("autodoc", "1");
+
+%{
+#include "mfem.hpp"
+#include "numpy/arrayobject.h"
+#include "../common/io_stream.hpp"
+#include "../common/pyoperator.hpp"
+#include "../common/pycoefficient.hpp"
+#include "../common/pyintrules.hpp"
+#include "../common/pynonlininteg.hpp"
+%}
+
+%include "../common/existing_mfem_headers.i"
+#ifdef FILE_EXISTS_FEM_HYPERBOLIC
+
+%init %{
+import_array();
+%}
+
+%include "exception.i"
+%include "std_string.i"
+%include "../common/exception.i"
+
+%import "array.i"
+%import "vector.i"
+%import "densemat.i"
+%import "eltrans.i"
+%import "nonlininteg.i"
+
+%include "fem/hyperbolic.hpp"
+
+#endif
+
+
+
diff --git a/mfem/_ser/lininteg.i b/mfem/_ser/lininteg.i
index bba39695e..428d0e6c9 100644
--- a/mfem/_ser/lininteg.i
+++ b/mfem/_ser/lininteg.i
@@ -19,10 +19,12 @@
import_array();
%}
+
+
%include "exception.i"
%import "globals.i"
-//%include "fem/coefficient.hpp"
+
%import "fe.i"
%import "vector.i"
%import "eltrans.i"
diff --git a/mfem/_ser/mesh.i b/mfem/_ser/mesh.i
index 8c2759cd7..d9c51a515 100644
--- a/mfem/_ser/mesh.i
+++ b/mfem/_ser/mesh.i
@@ -31,6 +31,7 @@ import_array();
%import "matrix.i"
%import "mem_manager.i"
%import "array.i"
+%import "attribute_sets.i"
%import "sort_pairs.i"
%import "ncmesh.i"
%import "vector.i"
@@ -222,30 +223,34 @@ def GetFaceElements(self, Face):
return Elem1.value(), Elem2.value()
%}
%feature("shadow") mfem::Mesh::GetElementTransformation %{
-def GetElementTransformation(self, i):
+def GetElementTransformation(self, i, Tr=None):
from mfem.ser import IsoparametricTransformation
- Tr = IsoparametricTransformation()
+ if Tr is None:
+ Tr = IsoparametricTransformation()
$action(self, i, Tr)
return Tr
%}
%feature("shadow") mfem::Mesh::GetBdrElementTransformation %{
-def GetBdrElementTransformation(self, i):
+def GetBdrElementTransformation(self, i, Tr=None):
from mfem.ser import IsoparametricTransformation
- Tr = IsoparametricTransformation()
+ if Tr is None:
+ Tr = IsoparametricTransformation()
$action(self, i, Tr)
return Tr
%}
%feature("shadow") mfem::Mesh::GetFaceTransformation %{
-def GetFaceTransformation(self, i):
+def GetFaceTransformation(self, i, Tr=None):
from mfem.ser import IsoparametricTransformation
- Tr = IsoparametricTransformation()
+ if Tr is None:
+ Tr = IsoparametricTransformation()
$action(self, i, Tr)
return Tr
%}
%feature("shadow") mfem::Mesh::GetEdgeTransformation %{
-def GetEdgeTransformation(self, i):
+def GetEdgeTransformation(self, i, Tr=None):
from mfem.ser import IsoparametricTransformation
- Tr = IsoparametricTransformation()
+ if Tr is None:
+ Tr = IsoparametricTransformation()
$action(self, i, Tr)
return Tr
%}
@@ -308,6 +313,10 @@ def CartesianPartitioning(self, nxyz, return_list=False):
%newobject mfem::Mesh::GetFaceToElementTable;
%newobject mfem::Mesh::GetVertexToElementTable;
+%immutable mfem::MeshPart::mesh;
+%immutable mfem::MeshPart::nodal_fes;
+%immutable mfem::MeshPart::nodes;
+
%include "../common/exception.i"
%include "mesh/mesh.hpp"
diff --git a/mfem/_ser/ncmesh.i b/mfem/_ser/ncmesh.i
index b0117c366..fd6158c93 100644
--- a/mfem/_ser/ncmesh.i
+++ b/mfem/_ser/ncmesh.i
@@ -36,6 +36,11 @@ XXXPTR_SIZE_IN(mfem::Refinement *data_, int asize, mfem::Refinement)
%immutable embeddings;
+%ignore mfem::NCMesh::FindFaceNodes(const Face &fa) const;
+%ignore mfem::NCMesh::GetElement(int i) const;
+%ignore mfem::NCMesh::GetFace(int i) const;
+%ignore mfem::NCMesh::GetNode(int i) const;
+
%include "mesh/ncmesh.hpp"
#ifndef SWIGIMPORTED
diff --git a/mfem/_ser/quadinterpolator.i b/mfem/_ser/quadinterpolator.i
index 9813ea3b9..48f79612b 100644
--- a/mfem/_ser/quadinterpolator.i
+++ b/mfem/_ser/quadinterpolator.i
@@ -25,6 +25,7 @@ import_array();
%import "../common/numpy_int_typemap.i"
+%include "../common/kernel_dispatch.i"
%include "fem/quadinterpolator.hpp"
#endif
diff --git a/mfem/_ser/setup.py b/mfem/_ser/setup.py
index ab14791f5..dbc6f6fdf 100644
--- a/mfem/_ser/setup.py
+++ b/mfem/_ser/setup.py
@@ -10,6 +10,8 @@
import os
import numpy
+# this remove *.py in this directory to be imported from setuptools
+sys.path.remove(os.path.abspath(os.path.dirname(sys.argv[0])))
from distutils.core import Extension, setup
@@ -104,8 +106,11 @@ def get_extensions():
"quadinterpolator", "quadinterpolator_face",
"submesh", "transfermap", "staticcond",
"sidredatacollection", "enzyme",
+ "attribute_sets", "arrays_by_name",
+ "hyperbolic",
"complex_densemat", "complexstaticcond", "complexweakform"]
+
if add_cuda == '1':
from setup_local import cudainc
include_dirs.append(cudainc)
diff --git a/mfem/_ser/solvers.i b/mfem/_ser/solvers.i
index e538bf391..507bf94c6 100644
--- a/mfem/_ser/solvers.i
+++ b/mfem/_ser/solvers.i
@@ -26,10 +26,17 @@ import_array();
%import "../common/operator_ptr_typemap.i"
%import "../common/exception_director.i"
-%ignore mfem::IterativeSolverMonitor::SetIterativeSolver;
%feature("director") mfem::IterativeSolverMonitor;
+%feature("director") mfem::IterativeSolverController;
%feature("director") mfem::PyIterativeSolver;
+
+// Forward declaration
+%inline %{
+ namespace mfem{
+ class IterativeSolver;
+ }
+%}
%include "linalg/solvers.hpp"
%include "../common/pysolvers.hpp"
diff --git a/mfem/_ser/sparsemat.i b/mfem/_ser/sparsemat.i
index 680404a9d..c80bb2cfa 100644
--- a/mfem/_ser/sparsemat.i
+++ b/mfem/_ser/sparsemat.i
@@ -99,7 +99,7 @@ if len(args) == 1 and isinstance(args[0], csr_matrix):
SparseMatrix(int *i, int *j, double *data, int m, int n);
allows to use numpy array to call this
*/
-%typemap(in) (int *i, int *j, double *data, int m, int n)
+%typemap(in) (int *i, int *j, mfem::real_t *data, int m, int n)
(PyArrayObject *tmp_arr1_ = NULL,
PyArrayObject *tmp_arr2_ = NULL,
PyArrayObject *tmp_arr3_ = NULL,
@@ -120,13 +120,13 @@ if len(args) == 1 and isinstance(args[0], csr_matrix):
//
PyArray_CLEARFLAGS(tmp_arr3_, NPY_ARRAY_OWNDATA);
}
-%typemap(freearg) (int *i, int *j, double *data, int m, int n){
+%typemap(freearg) (int *i, int *j, mfem::real_t *data, int m, int n){
//Py_XDECREF(tmp_arr1_$argnum); Dont do this.. We set OwnsGraph and OwnsData to Fase in Python
//Py_XDECREF(tmp_arr2_$argnum);
//Py_XDECREF(tmp_arr3_$argnum);
}
-%typemap(typecheck ) (int *i, int *j, double *data, int m, int n){
+%typemap(typecheck ) (int *i, int *j, mfem::real_t *data, int m, int n){
/* check if list of 5 numpy array or not */
if (!PyList_Check($input)) $1 = 0;
else {
diff --git a/mfem/_ser/transfer.i b/mfem/_ser/transfer.i
index b243bbd86..10955b7ec 100644
--- a/mfem/_ser/transfer.i
+++ b/mfem/_ser/transfer.i
@@ -17,6 +17,7 @@ import_array();
%include "exception.i"
%import "operators.i"
+%import "device.i"
%import "fespace.i"
%include "../common/exception.i"
diff --git a/mfem/common/array_instantiation_macro.i b/mfem/common/array_instantiation_macro.i
index aac077192..7d4d46278 100644
--- a/mfem/common/array_instantiation_macro.i
+++ b/mfem/common/array_instantiation_macro.i
@@ -1,6 +1,7 @@
%define INSTANTIATE_ARRAY2(XXX, YYY, ZZZ, USEPTR)
#if USEPTR == 1
-%template(##ZZZ##Ptr##Array) mfem::Array;
+ //%template(##ZZZ##Ptr##Array) mfem::Array;
+%template(##ZZZ##Array) mfem::Array;
#else
%template(##ZZZ##Array) mfem::Array;
#endif
@@ -325,6 +326,8 @@ INSTANTIATE_ARRAY2(XXX, YYY, YYY, USEPTR)
%ignore mfem::Array2D::Print;
%ignore mfem::Array2D::PrintGZ;
%ignore mfem::Array2D::SaveGZ;
+%ignore mfem::Array2D::Load;
+%ignore mfem::Array2D::Save;
%enddef
diff --git a/mfem/common/array_listtuple_typemap.i b/mfem/common/array_listtuple_typemap.i
index 9b263fca9..4bbda0499 100644
--- a/mfem/common/array_listtuple_typemap.i
+++ b/mfem/common/array_listtuple_typemap.i
@@ -71,6 +71,7 @@
if (!SWIG_IsOK(SWIG_ConvertPtr(s, (void **) &temp_ptr$argnum, ty, 0|0))) {
PyErr_SetString(PyExc_ValueError, "List items must be XXX");
} else {
+ PyObject_SetAttrString(s, "thisown", Py_False);
#if USEPTR==1
(* result)[i] = temp_ptr$argnum;
#else
diff --git a/mfem/common/bessel.py b/mfem/common/bessel.py
new file mode 100644
index 000000000..112c6afbd
--- /dev/null
+++ b/mfem/common/bessel.py
@@ -0,0 +1,335 @@
+'''
+ bessel functions (real order, complex argument)
+ iv
+ kv
+ jv
+ hankel1
+ hankel2
+ yv
+'''
+import numpy as np
+from numpy import exp, log, pi, sin, cos, sqrt, abs, cosh, sinh, sum
+from math import gamma
+from numba import njit, float64, int64, complex128
+
+# machine precision for double computation
+E = np.finfo(np.complex128).resolution # 1e-15
+D = -np.log10(np.finfo(np.complex128).resolution) # 15
+
+'''
+I(n, x)
+ n: real
+ x: compplex
+'''
+
+
+def iv_smallz(n, z):
+ '''
+ |z| < 2*sqrt(n+1)
+ '''
+ a = 1
+ sum1 = 0
+ for k in range(20):
+ sum1 = sum1 + a
+ fac = 1/(k+1)/(k+n+1)*(z**2/4)
+ a = a*fac
+ if abs(fac) < E:
+ break
+
+ return sum1*(z/2)**n/gamma(1+n)
+
+
+def iv_largez(n, z):
+ '''
+ good for |z| > max(1.2*D+2.4, n**2/2)
+ '''
+ a = -(4*n**2 - 1)/(8*z)
+ b = (4*n**2 - 1)/(8*z)
+
+ sum1 = 1
+ sum2 = 1
+ for i in range(15):
+ k = i+1
+ sum1 = sum1 + a
+ sum2 = sum2 + b
+ k = k+1
+ a = -a*(4*n**2 - (2*k-1)**2)/k/(8*z)
+ b = b*(4*n**2 - (2*k-1)**2)/k/(8*z)
+
+ ans = exp(z)/sqrt(2*pi*z) * sum1
+ if np.imag(z) >= 0:
+ ans = ans + sum2*exp(1j*(n+0.5)*pi)*exp(-z)/sqrt(2*pi*z)
+ else:
+ ans = ans + sum2*exp(-1j*(n+0.5)*pi)*exp(-z)/sqrt(2*pi*z)
+
+ return ans
+
+
+def iv_middlez(n, z):
+ nu = n - np.floor(n)
+ size = 40
+ data = np.zeros(size, dtype=np.complex128)
+ data[-2] = 1
+
+ for i in range(len(data)-2):
+ nn = len(data)-2-i+nu
+ data[-3-i] = 2*nn/z*data[-2-i] + data[-1-i]
+
+ norm = data[0]
+ for k in range(len(data)):
+ if k == 0:
+ norm = data[0]
+ fac = 1.
+ else:
+ fac = fac*k
+ lam = 2*(nu+k)*gamma(k+2*nu)/gamma(1+2*nu)/fac
+ norm += data[k]*lam
+
+ data /= norm
+ data *= (z/2.)**nu/gamma(1+nu)*exp(z)
+
+ return data[int(np.floor(n))]
+
+
+def kv_smallz(n, z):
+ '''
+ |z| > 2, np.real(z) > 0
+ '''
+ tmp = n-np.floor(n)
+ nu = tmp if tmp < 0.5 else tmp-1.
+
+ if nu == -0.5:
+ k1 = exp(-z)*sqrt(pi/2/z)
+ k2 = exp(-z)*sqrt(pi/2/z)
+
+ if nu != 0:
+ g1 = (1./gamma(1-nu) - 1./gamma(1+nu))/(2*nu)
+ else:
+ nu1 = 1e-5
+ g1 = (1/gamma(1-nu1) - 1/gamma(1+nu1))/(2.*nu1)
+
+ g2 = (1/gamma(1-nu) + 1/gamma(1+nu))/2.0
+
+ aa = gamma(1+nu)*gamma(1-nu)
+
+ if abs(nu) == 0.0:
+ term1 = 1.0
+ term2 = 1.0
+ else:
+ mu = nu*log(2/z)
+ term1 = cosh(mu)
+ term2 = sinh(mu)/mu
+
+ fk = aa*(g1*term1 + g2*log(2./z)*term2)
+ pk = 0.5*(z/2.)**(-nu)*gamma(1+nu)
+ qk = 0.5*(z/2.)**(nu)*gamma(1-nu)
+ ck = 1.
+
+ k1 = ck*fk
+ k2 = ck*pk
+ for i in range(30):
+ k = i+1
+
+ fk = (k*fk + pk + qk)/(k**2-nu**2)
+
+ pk = pk/(k-nu)
+ qk = qk/(k+nu)
+ ck = ck*(z**2/4)/k
+
+ k1 = k1 + ck*fk
+ k2 = k2 + ck*(pk - k*fk)
+
+ k2 = 2/z*k2
+
+ num_rec = int(n - nu)
+
+ if num_rec == 0:
+ return k1
+ if num_rec == 1:
+ return k2
+
+ for i in range(num_rec-1):
+ nn = i+1+nu
+ tmp = k2
+ k2 = 2*nn/z*k2+k1
+ k1 = tmp
+
+ return k2
+
+
+def kv_largez(n, z):
+ '''
+ |z| > 2, np.real(z) > 0
+ '''
+ tmp = n-np.floor(n)
+ nu = tmp if tmp < 0.5 else tmp-1.
+
+ if nu == -0.5:
+ k1 = exp(-z)*sqrt(pi/2/z)
+ k2 = k1
+ else:
+ size = 50
+ data = np.zeros(size, np.complex128)
+ data[1] = 1.
+
+ for i in range(size-2):
+ nn = size-i-2.
+
+ an = ((nn-0.5)**2 - nu**2)/nn/(nn+1)
+ bn = 2*(nn+z)/(nn+1)
+
+ data[i+2] = -(data[i] - bn*data[i+1])/an
+
+ data /= sum(data)
+ data *= (2*z)**(-nu-0.5)
+
+ fac = cos(pi*nu)/pi*gamma(nu+0.5)*gamma(0.5-nu)
+ uu0 = data[-1]/fac
+
+ k1 = sqrt(pi)*(2*z)**nu * exp(-z)*uu0
+ k2 = k1*(nu+0.5+z - data[-2]/data[-1])/z
+
+ num_rec = int(n - nu)
+
+ if num_rec == 0:
+ return k1
+ if num_rec == 1:
+ return k2
+
+ for i in range(num_rec-1):
+ nn = i+1+nu
+ tmp = k2
+ k2 = 2*nn/z*k2+k1
+ k1 = tmp
+
+ return k2
+
+
+def kv_righthalf(n, z):
+ if abs(z) < 2:
+ kval = kv_smallz(abs(n), z)
+ else:
+ kval = kv_largez(abs(n), z)
+ return kval
+
+
+def iv_righthalf(n, z):
+ safe_guard = 1.0 # must be >1, expand intermidate region
+ if abs(z) < 2*sqrt(abs(n)+1)/safe_guard:
+ ival = iv_smallz(abs(n), z)
+ elif abs(z) > (1.2*D+2.4)*safe_guard and abs(z) > (abs(n)**2/2)*safe_guard:
+ ival = iv_largez(abs(n), z)
+ else:
+ ival = iv_middlez(abs(n), z)
+
+ if n < 0:
+ kval = kv_righthalf(n, z)
+ ival = ival + 2./pi*sin(-n*pi)*kval
+
+ if np.imag(z) == 0:
+ return np.real(ival)
+
+ return ival
+
+
+def jv_righthalf(n, z):
+ jval = exp(1j*n*pi/2.)*iv_righthalf(n, z/1j)
+ if np.imag(z) == 0:
+ return np.real(jval)
+ return jval
+
+#
+# kv, iv, jv, hankel1, hankel2, yv
+#
+
+
+def kv(n, z):
+ if np.real(z) < 0:
+ zz = -z
+ ival = iv_righthalf(n, zz)
+ else:
+ zz = z
+ ival = 0j
+
+ kval = kv_righthalf(n, zz)
+
+ ee = exp(1j*pi*n)
+ if (n-np.floor(n)) == 0.0:
+ ee = np.real(ee)
+
+ if np.real(z) < 0 and np.imag(z) >= 0:
+ return 1/ee*kval - 1j*pi*ival
+ elif np.real(z) < 0 and np.imag(z) < 0:
+ return ee*kval + 1j*pi*ival
+ else:
+ return kval
+
+
+def iv(n, z):
+ if np.real(z) < 0:
+ zz = -z
+ else:
+ zz = z
+ ival = iv_righthalf(n, zz)
+
+ ee = exp(1j*pi*n)
+ if (n-np.floor(n)) == 0.0:
+ ee = np.real(ee)
+
+ if -np.real(z) > 0 and np.imag(z) >= 0:
+ return ee*ival
+ elif -np.real(z) > 0 and np.imag(z) < 0:
+ return 1/ee*ival
+
+ return ival
+
+
+def jv(n, z):
+ if -np.real(z) > 0:
+ zz = -z
+ else:
+ zz = z
+ jval = jv_righthalf(n, zz)
+
+ ee = exp(1j*pi*n)
+ if (n-np.floor(n)) == 0.0:
+ ee = np.real(ee)
+
+ if -np.real(z) > 0 and np.imag(z) >= 0:
+ return ee*jval
+ elif -np.real(z) > 0 and np.imag(z) < 0:
+ return 1/ee*jval
+
+ return jval
+
+
+def hankel1(n, z):
+ zz = z*exp(-1j*pi/2)
+ return 2/pi/1j*exp(-1j*pi*n/2)*kv(n, zz)
+
+
+def hankel2(n, z):
+ zz = z*exp(1j*pi/2)
+ return -2/pi/1j*exp(1j*pi*n/2)*kv(n, zz)
+
+
+def yv(n, z):
+ return (hankel1(n, z) - hankel2(n, z))/2/1j
+
+
+jitter = njit(complex128(float64, complex128))
+
+iv_smallz = jitter(iv_smallz)
+iv_largez = jitter(iv_largez)
+iv_middlez = jitter(iv_middlez)
+kv_smallz = jitter(kv_smallz)
+kv_largez = jitter(kv_largez)
+kv_righthalf = jitter(kv_righthalf)
+iv_righthalf = jitter(iv_righthalf)
+jv_righthalf = jitter(jv_righthalf)
+kv = jitter(kv)
+iv = jitter(iv)
+jv = jitter(jv)
+hankel1 = jitter(hankel1)
+hankel2 = jitter(hankel2)
+yv = jitter(yv)
diff --git a/mfem/common/bilininteg_ext.i b/mfem/common/bilininteg_ext.i
index 99821135c..c6b093ce5 100644
--- a/mfem/common/bilininteg_ext.i
+++ b/mfem/common/bilininteg_ext.i
@@ -219,6 +219,10 @@ namespace mfem {
self._coeff = args
%}
+%pythonappend VectorFEBoundaryFluxIntegrator::VectorFEBoundaryFluxIntegrator %{
+ self._coeff = args
+%}
+
%pythonappend DerivativeIntegrator::DerivativeIntegrator %{
self._coeff = q
%}
diff --git a/mfem/common/generate_bilininteg_ext.py b/mfem/common/generate_bilininteg_ext.py
index 84229e9df..06d882700 100644
--- a/mfem/common/generate_bilininteg_ext.py
+++ b/mfem/common/generate_bilininteg_ext.py
@@ -36,6 +36,10 @@
pass
elif line.find("(self, fes, e=1.0)") != -1:
pass
+ elif line.find("(self, vdim_=1)") != -1:
+ pass
+ elif line.find("(self, vdim_)") != -1:
+ pass
else:
print(cname)
print(line)
diff --git a/mfem/common/generate_lininteg_ext.py b/mfem/common/generate_lininteg_ext.py
index a8c943e48..6d3c272d8 100644
--- a/mfem/common/generate_lininteg_ext.py
+++ b/mfem/common/generate_lininteg_ext.py
@@ -22,6 +22,8 @@
pp = " self._coeff = QF"
elif line.find(", F)") != -1:
pp = " self._coeff = F"
+ elif line.find(", f)") != -1:
+ pp = " self._coeff = f"
elif line.find(", f, s=1.0, ir=None)") != -1:
pp = " self._coeff = (f, ir)"
elif line.find(", uD_, lambda_, mu_, alpha_, kappa_)") != -1:
diff --git a/mfem/common/kernel_dispatch.i b/mfem/common/kernel_dispatch.i
new file mode 100644
index 000000000..20475486a
--- /dev/null
+++ b/mfem/common/kernel_dispatch.i
@@ -0,0 +1,6 @@
+/*
+ kernel_dispatch.i
+
+ We set a macro simpily ingnore the MFEM_REGISTER_KERNELS macro
+ */
+#define MFEM_REGISTER_KERNELS(...) //
diff --git a/mfem/common/lininteg_ext.i b/mfem/common/lininteg_ext.i
index 4c3643fc7..ebd59a46e 100644
--- a/mfem/common/lininteg_ext.i
+++ b/mfem/common/lininteg_ext.i
@@ -44,6 +44,9 @@ namespace mfem {
%pythonappend VectorFEBoundaryFluxLFIntegrator::VectorFEBoundaryFluxLFIntegrator %{
self._coeff = args
%}
+%pythonappend VectorFEBoundaryNormalLFIntegrator::VectorFEBoundaryNormalLFIntegrator %{
+ self._coeff = f
+%}
%pythonappend VectorFEBoundaryTangentLFIntegrator::VectorFEBoundaryTangentLFIntegrator %{
self._coeff = QG
%}
diff --git a/mfem/par.py b/mfem/par.py
index 82ea4d50a..4bc260d12 100644
--- a/mfem/par.py
+++ b/mfem/par.py
@@ -77,6 +77,7 @@
from mfem._par.qfunction import *
from mfem._par.quadinterpolator import *
from mfem._par.quadinterpolator_face import *
+from mfem._par.attribute_sets import *
from mfem._par.fe_base import *
from mfem._par.fe_h1 import *
@@ -93,6 +94,7 @@
from mfem._par.psubmesh import *
from mfem._par.transfermap import *
from mfem._par.ptransfermap import *
+from mfem._par.hyperbolic import *
try:
from mfem._par.gslib import *
diff --git a/mfem/ser.py b/mfem/ser.py
index c57456369..24eaa32bc 100644
--- a/mfem/ser.py
+++ b/mfem/ser.py
@@ -60,6 +60,7 @@
from mfem._ser.qfunction import *
from mfem._ser.quadinterpolator import *
from mfem._ser.quadinterpolator_face import *
+from mfem._ser.attribute_sets import *
from mfem._ser.fe_base import *
from mfem._ser.fe_h1 import *
@@ -72,10 +73,15 @@
from mfem._ser.fe_nurbs import *
from mfem._ser.doftrans import *
from mfem._ser.std_vectors import *
-from mfem._ser.complex_densemat import *
+
+try:
+ from mfem._ser.complex_densemat import *
+except ImportError:
+ pass
from mfem._ser.submesh import *
from mfem._ser.transfermap import *
+from mfem._ser.hyperbolic import *
import mfem._ser.array as array
import mfem._ser.blockoperator as blockoperator
diff --git a/requirements.txt b/requirements.txt
index 5d422a6c6..88fe13801 100644
--- a/requirements.txt
+++ b/requirements.txt
@@ -1,6 +1,5 @@
-numpy>=1.19.4; python_version<"3.7"
-numpy>=1.20.0; python_version>="3.7"
+numpy >= 1.22.4
+numba
scipy
-swig >= 4.1.1
+swig >= 4.3
cmake
-
diff --git a/setup.py b/setup.py
index e603d24da..b64d4c93e 100644
--- a/setup.py
+++ b/setup.py
@@ -50,7 +50,7 @@
# "metis": "http://glaros.dtc.umn.edu/gkhome/fetch/sw/metis/metis-5.1.0.tar.gz",
"metis": "https://github.com/mfem/tpls/raw/gh-pages/metis-5.1.0.tar.gz",
"hypre": "https://github.com/hypre-space/hypre/archive/v2.28.0.tar.gz",
- "libceed": "https://github.com/CEED/libCEED/archive/refs/tags/v0.11.0.tar.gz",
+ "libceed": "https://github.com/CEED/libCEED/archive/refs/tags/v0.12.0.tar.gz",
"gslib": "https://github.com/Nek5000/gslib/archive/refs/tags/v1.0.8.tar.gz"}
repos = {"mfem": "https://github.com/mfem/mfem.git",
@@ -59,12 +59,8 @@
"metis": "https://github.com/KarypisLab/METIS", }
repos_sha = {
- # "mfem": "00b2a0705f647e17a1d4ffcb289adca503f28d42", # version 4.5.2
- # "mfem": "962774d5ffa84ceed3bc670e52388250ee028da1", # version 4.5.2 + distsolve
- # "mfem": "69fbae732d5279c8d0f42c5430c4fd5656731d00", # version 4.6
- # "mfem": "8bb929c2ff86cdf2ee9bb058cc75e59acb07bb94", # doftrans simplification (Nov. 15. 2023)
- # "mfem": "4a45c70d1269d293266b77a3a025a9756d10ed8f", # socket connection fix (Nov. 29 2023)
- "mfem": "68873fa4d403c7c94a653c7bc781815ff5b2734d", # use constrained prolongation operator in ex26, ex26p (Nov. 30 2023)
+ "mfem": "a01719101027383954b69af1777dc828bf795d62", # v4.8
+ # "mfem": "dc9128ef596e84daf1138aa3046b826bba9d259f", # v4.7
"gklib": "a7f8172703cf6e999dd0710eb279bba513da4fec",
"metis": "94c03a6e2d1860128c2d0675cbbb86ad4f261256", }
@@ -217,14 +213,14 @@ def read_mfem_tplflags(prefix):
'long_description_content_type': "text/markdown",
'url': 'http://mfem.org',
'download_url': 'https://github.com/mfem',
- 'classifiers': ['Development Status :: 4 - Beta',
+ 'classifiers': ['Development Status :: 5 - Production/Stable',
'Intended Audience :: Developers',
'Topic :: Scientific/Engineering :: Physics',
'License :: OSI Approved :: BSD License',
- 'Programming Language :: Python :: 3.7',
'Programming Language :: Python :: 3.8',
'Programming Language :: Python :: 3.9',
- 'Programming Language :: Python :: 3.10', ],
+ 'Programming Language :: Python :: 3.10',
+ 'Programming Language :: Python :: 3.11', ],
'keywords': [k for k in keywords.split('\n') if k],
'platforms': [p for p in platforms.split('\n') if p],
'license': 'BSD-3',
@@ -303,7 +299,7 @@ def find_command(name):
assert False, "SWIG is not installed (hint: pip install swig)"
-def make_call(command, target='', force_verbose=False):
+def make_call(command, target='', force_verbose=False, env=None):
'''
call command
'''
@@ -311,7 +307,9 @@ def make_call(command, target='', force_verbose=False):
if dry_run:
return
- kwargs = {'universal_newlines': True}
+ kwargs = {'universal_newlines': True, 'env': env}
+ if env is not None:
+ env.update(os.environ)
myverbose = verbose or force_verbose
if not myverbose:
@@ -673,7 +671,7 @@ def make_metis(use_int64=False, use_real64=False):
command = ['make', 'config', 'shared=1',
'prefix=' + metis_prefix,
'cc=' + cc_command]
- make_call(command)
+ make_call(command, env={'CMAKE_POLICY_VERSION_MINIMUM': '3.5'})
make('metis')
make_install('metis')
diff --git a/test/test_blockmatrix.py b/test/test_blockmatrix.py
index c78066ba7..fe29711ce 100644
--- a/test/test_blockmatrix.py
+++ b/test/test_blockmatrix.py
@@ -35,3 +35,7 @@
m.SetBlock(1, 0, mmat)
print(m._offsets)
print(m._linked_mat)
+
+from mfem.common.sparse_utils import sparsemat_to_scipycsr
+print(m.CreateMonolithic())
+print(sparsemat_to_scipycsr(m.CreateMonolithic(), np.float64).todense())
diff --git a/test/test_coefficient.py b/test/test_coefficient.py
new file mode 100644
index 000000000..65204227e
--- /dev/null
+++ b/test/test_coefficient.py
@@ -0,0 +1,67 @@
+import sys
+import numpy as np
+import gc
+from scipy.sparse import csr_matrix
+
+if len(sys.argv) > 1 and sys.argv[1] == '-p':
+ import mfem.par as mfem
+ use_parallel = True
+
+ from mfem.common.mpi_debug import nicePrint as print
+
+ from mpi4py import MPI
+ myid = MPI.COMM_WORLD.rank
+
+else:
+ import mfem.ser as mfem
+ use_parallel = False
+ myid = 0
+
+def test():
+ dim = 3
+ max_attr = 5
+ sigma_attr_coefs = mfem.MatrixCoefficientArray(max_attr)
+ sigma_attr = mfem.intArray(max_attr)
+
+
+ tensors = [mfem.DenseMatrix(np.ones((3,3))*i) for i in range(max_attr)]
+ tensor_coefs = [mfem.MatrixConstantCoefficient(mat) for mat in tensors]
+ sigma_array = mfem.MatrixCoefficientArray(tensor_coefs)
+ sigma_attr = mfem.intArray([1,2,3,4,5])
+ sigmaCoef = mfem.PWMatrixCoefficient(dim, sigma_attr, sigma_array, False)
+
+ for ti, tensor in enumerate(tensors):
+ # add matrix coefficient to list
+ xx = mfem.MatrixConstantCoefficient(tensor)
+ sigma_attr_coefs[ti] = xx
+ sigma_attr[ti] = ti+1
+
+ # Create PW Matrix Coefficient
+ sigmaCoef = mfem.PWMatrixCoefficient(dim, sigma_attr, sigma_attr_coefs, False)
+
+ sigmaCoef = mfem.PWMatrixCoefficient(dim, sigma_attr, tensor_coefs, False)
+ tensor_coefs = mfem.MatrixCoefficientArray([mfem.MatrixConstantCoefficient(mat) for mat in tensors])
+ sigmaCoef = mfem.PWMatrixCoefficient(dim, sigma_attr, tensor_coefs, False)
+
+ data = tensor_coefs.GetData()
+ tensor_coefs2 = mfem.MatrixCoefficientArray(data, 5, False)
+ sigmaCoef = mfem.PWMatrixCoefficient(dim, sigma_attr, tensor_coefs2, False)
+
+ print("exiting")
+
+
+if __name__ == '__main__':
+ import tracemalloc
+
+ tracemalloc.start()
+
+ print(tracemalloc.get_traced_memory())
+
+ for i in range(3000):
+ test()
+ print(tracemalloc.get_traced_memory())
+ snapshot = tracemalloc.take_snapshot()
+ top_stats = snapshot.statistics('lineno')
+ for stat in top_stats[:10]:
+ print(stat)
+
diff --git a/test/test_intrules.py b/test/test_intrules.py
index 542d027eb..a1692fc25 100644
--- a/test/test_intrules.py
+++ b/test/test_intrules.py
@@ -52,13 +52,13 @@ def run_test():
irs = [mfem.IntRules.Get(i, 2) for i in range(mfem.Geometry.NumGeom)]
- rulearray = mfem.IntegrationRulePtrArray(irs)
+ rulearray = mfem.IntegrationRuleArray(irs)
ir2 = rulearray[2]
points3 = [(ir2[i].x, ir2[i].y) for i in range(ir2.GetNPoints())]
assert (points3 == points), "IntegrationPointArray coords does not agree (check 2)."
- # check slice of IntegrationRulePtrArray
+ # check slice of IntegrationRuleArray
rulearray2 = rulearray[:5]
assert not rulearray2.OwnsData(), "subarray should not own data"
ir2 = rulearray2[2]
@@ -67,7 +67,7 @@ def run_test():
# create it from a pointer array
data = rulearray2.GetData() # this returns
- rulearray3 = mfem.IntegrationRulePtrArray((data, 5))
+ rulearray3 = mfem.IntegrationRuleArray((data, 5))
ir2 = rulearray3[2]
points3 = [(ir2[i].x, ir2[i].y) for i in range(ir2.GetNPoints())]
assert (points3 == points), "IntegrationPointArray coords does not agree (check 3)."
diff --git a/test/test_vector.py b/test/test_vector.py
index 3fc69f98c..99ab1c3fb 100644
--- a/test/test_vector.py
+++ b/test/test_vector.py
@@ -29,7 +29,7 @@ def run_test():
a.Print_HYPRE("vector_hypre.dat")
- x = mfem.VectorPtrArray([a]*3)
+ x = mfem.VectorArray([a]*3)
x[2].Print()
if __name__=='__main__':